Lab Notes
Short stories and links shared by the scientists in our community
Scientists find the first known nursery for the Munk's devil ray
By tracking rays across the eastern Pacific, researchers spied the growth and development of this less common ray species
David Sim on Wikimedia Commons (CC BY 2.0)
Munk's devil rays (Mobula munkiana) are endemic to the Eastern Pacific, especially around the Gulf of California in Mexico. Despite their common sightings, much of their behavior remains a mystery for scientists. Being in a vulnerable conservation state, finding out more about them and their reproductive habits, could prove crucial for their protection.
These mobulas are renowned for their social behavior, gathering in big groups normally by size. During the past years, local fishermen and tourism operators had observed consistent aggregations of these rays in Ensenada Grande, a shallow bay in the Espiritu Santo Archipelago in the Gulf of California. A research team focused on this area, aiming to find out if a particular age group was predominant in these gatherings.
The team captured 95 rays in the bay during different periods, measured and classified them by their physical features as neonates, juveniles, or adults. Using conventional tagging and acoustic telemetry, they sampled their location and followed their movements for 22 months. The captured rays were mainly juveniles and neonates, and their tracking confirmed that the individuals stayed in Ensenada Grande for several months.
To find out if the juveniles and neonates appeared at the same time every year in this location, the scientists used professional photographs taken in recent years, which confirmed this hypothesis. These data lead to the conclusion that young mobulas use this shallow and protected bay as a nursery, where they grow until becoming less vulnerable.
Thanks to this study, the first nursery area for the Mobula munkiana has been discovered. Being mobulas under threat by targeted or accidental fishing, this finding could be vital for their conservation. By protecting this area, we could make sure their rare offspring has better chances of survival.
What is a comet? Your answer may depend on the language you learn in
Some astronomical terms get lost in translation, depending on the language
Maël Balland on Pexels.
Astronomy, one of the oldest sciences, is awe-inspiring. Studying astronomy can be as simple as peering out at the universe through a telescope, watching a meteor shower, or simply looking out your window at night at the stars. But, can we really define the concept of the universe? If we ask an adult that question, we'll probably get a mostly correct answer. But when we ask the same question to children, the answer might surprise us, depending on what language they speak.
In a study published in Participatory Educational Research, more than 100 students from 7th grade (11 to 13 years old) were asked about their concepts of the universe, the Sun, comets, and constellations. They were given four options, only one of which was scientifically correct. The other ones represented misconceptions in astronomy, which children are usually exposed to through everyday experiences such as incorrect concept formation in school, unscientific use of language, and even changes in meaning when words are translated between languages. These experiences can misinform students and that can affect their understandings of astronomy.
For example, when the children were asked to define a comet, these were the options presented:
- Comets are fast-moving stars
- Comets are non-moving stars
- Comets are not stars, they reflect the light of the sun and are made of frozen ice, dust, and gas
- A comet is a falling star, as the stars fall down, they appear to have a tail, and people make wishes when they see them
Only the third option was correct. Nonetheless, just 20 percent of all participants could answer and explain their option correctly. The researchers used these findings to discuss how the languages that children speak could influence their understanding of the concept: In Turkish, the term “comet” is translated as “tailed star”. Similarly, in Spanish, the term meteor shower is translated to “rain of stars”, when in reality meteor showers do not involve stars at all. This study suggests that a revision of the concepts given to children is necessary, especially when the language itself can bring misconceptions.
Lightning blasts on clay-rich soils support a dramatic theory of how life began on Earth
Phosphorus in clay-rich soils suggest that lightning was a key ingredient in life's recipe
Phosphorus is a key element in organic compounds, including the backbones of DNA and its cousin, RNA. But where did the phosphorus used in life's earliest chemical reactions come from? Scientists have proposed that meteorites could have a key source of reactive phosphorus, but a recent study illuminates the role lightning may have played in kick-starting life on prebiotic Earth.
When schreibersite, a mineral containing iron, nickel, and phosphorus, gets hydrated, it becomes capable of making organic molecules. Schreibersite has been a known source of this form of reactive phosphorus but until now, non-meteorite sources of schreibersite were thought to be extremely rare. Such sources include special forms of glass produced by lightning strikes, called fulgurites. Fulgurites can form when lightning strikes the ground and the extreme heat changes the material impacted by the strike.
In a new study published in Nature Communications, planetary scientists investigated minerals near a lightning strike site in Illinois. The researchers, from Yale University and the University of Leeds, found that a fulgurite produced in clay-rich soil contained a large amount of schreibersite. This finding raised the possibility that lightning strikes may have played an important role in creating the reactive phosphorus needed for life on Earth.
The researchers used the fulgurite from Illinois and previous reports of schreibersite in fulgurites to estimate the amount of reactive phosphorus produced from each lightning strike. They combined this information with a calculated annual lightning strike rate to estimate how much reactive phosphorus would have formed from lightning strikes during the Hadean and Archean eons.
The researchers estimated between 10 and 1000 kilograms of phosphide, and between 100 and 10,000 kilograms of other reactive forms of phosphorus (phosphite and hypophosphite) would have formed annually due to lightning strikes during these time periods. Their estimates rely on the current models of early Earth characteristics.
The findings of this study indicate lightning as a potentially meaningful source of terrestrial reactive phosphorus needed for life to start on Earth, suggesting there may be more than meteorites to our origins.
Some lucid dreamers can answer questions and math problems in their sleep
For the first time, scientists have been able to communicate with people while they are dreaming
Photo by Bruce Christianson on Unsplash
Lucid dreaming, or the ability to become aware that you're dreaming while you are actively doing so, is a rare psychological state that has long fascinated scientists. So far, scientists had been able to show how participants can process external cues while remaining asleep but never communicate back while asleep.
This has changed with the publication of a new research study in Current Biology detailing how scientists were able communicate in real-time with lucid dreamers instead of having to wait for study participants to wake up and talk about their dreams.
While sleeping, the volunteers communicated with the researchers, responded to questions, and answered math problems by moving their eyes left and right and contracting their facial muscles. For example, one study participant was asked to solve the math problem "four minus zero" and answered by moving their eyes left and right four times each — all while sleeping. Moreover, volunteers typically were able to recollect what happened upon awakening.
The possibility to hear the outside world during the stage of dreaming opens up a world of opportunities for some people including, but not limited to, interactive dreaming, recreational enjoyment or even lucid dreaming therapy.
Your brain responds to why you're drinking, not just what you're drinking
Drinking for relief produces a different reaction in the brain than drinking for reward does
Photo by Tomáš Malík on Unsplash
Different people drink alcohol for different reasons. Knowing someone’s motivation can help researchers develop more personalized treatments for problematic drinking. Studies show that reward drinkers, those who say that drinking makes them feel good, behave differently from relief drinkers, those who drink because it makes them feel less bad (i.e., they are self-medicating or alleviating withdrawal symptoms).
In a recent study, we aimed to explore whether these two categories of heavy drinkers also showed differences in their brains. Specifically, do reward and relief drinkers have different patterns of neural activation when looking at pictures of alcoholic beverages?
To answer this question, we recruited people who drink heavily and categorized them into reward- and relief-drinking groups using the Reward/Relief/Habit Drinking Scale and the Reasons for Heavy Drinking Questionnaire. Previous research suggested that while most people begin drinking for positive reinforcement (reward), as they continue to drink heavily for a long period of time, they begin to drink out of negative reinforcement (relief).
We also knew that the ventral striatum is a brain region associated with reward, while the dorsal striatum is associated with compulsive behavior. Therefore, we hypothesized that reward drinkers would have greater neural activation in the ventral striatum while looking at images of alcoholic drinks, whereas relief drinkers would have greater activation in the dorsal striatum when viewing these images.
We found that relief drinkers did indeed show significantly more activation in the dorsal striatum. However, contrary to our hypothesis, there was not much difference between reward and relief drinkers in the ventral striatum. We interpreted this to mean that the rewarding qualities of alcohol may not be lost in relief drinkers, but that a sense of relief may be gained in addition to the reward.
This study showed that there might be biological differences underlying different motivations for drinking, which opens the door to further development of precision medicine to treat alcohol addiction. One next step in this line of inquiry might be to examine differences in reward and relief drinkers’ response to existing treatments.
These are the health effects of tear gas
Tear gas can kill, but exposure can also cause chronic respiratory difficulty and severe eye injuries for months and potentially years after
Rose Pineda via Wikimedia
A tear gas canister contains a few different things. Primarily they are made of an irritant, most commonly 2-chlorobenzalmalononitrile (CS). Another irritant, 1‐chloroacetophenone (CN) was common until the 1980s, but it was found to not be potent or stable enough. A common composition for a complete tear gas cocktail is 45% CS, 30% potassium chlorate, 14% epoxy resin, 7% maleic anhydride, 3% methyl nadic anhydride, with some small residual ingredients. Although CS is the main cause of pain and irritation, all these ingredients have their own inherent toxicities. Potassium chlorate, for instance, cause burns and irritation itself, and can cause anemia if inhaled.
Tear gas agents like CS and CN function by interacting with TRPA1, a receptor protein expressed on the surface of nociceptors, pain-sensing neurons. In a way that's similar to how strong mustard makes your nose burn, tear gas agents bind to these proteins at minuscule concentrations, tens of thousands of times more potently than mustard, over-activating nociceptors. Tear gas causes extremely painful burning and irritation in the eyes, mouth, nose, and respiratory tract.
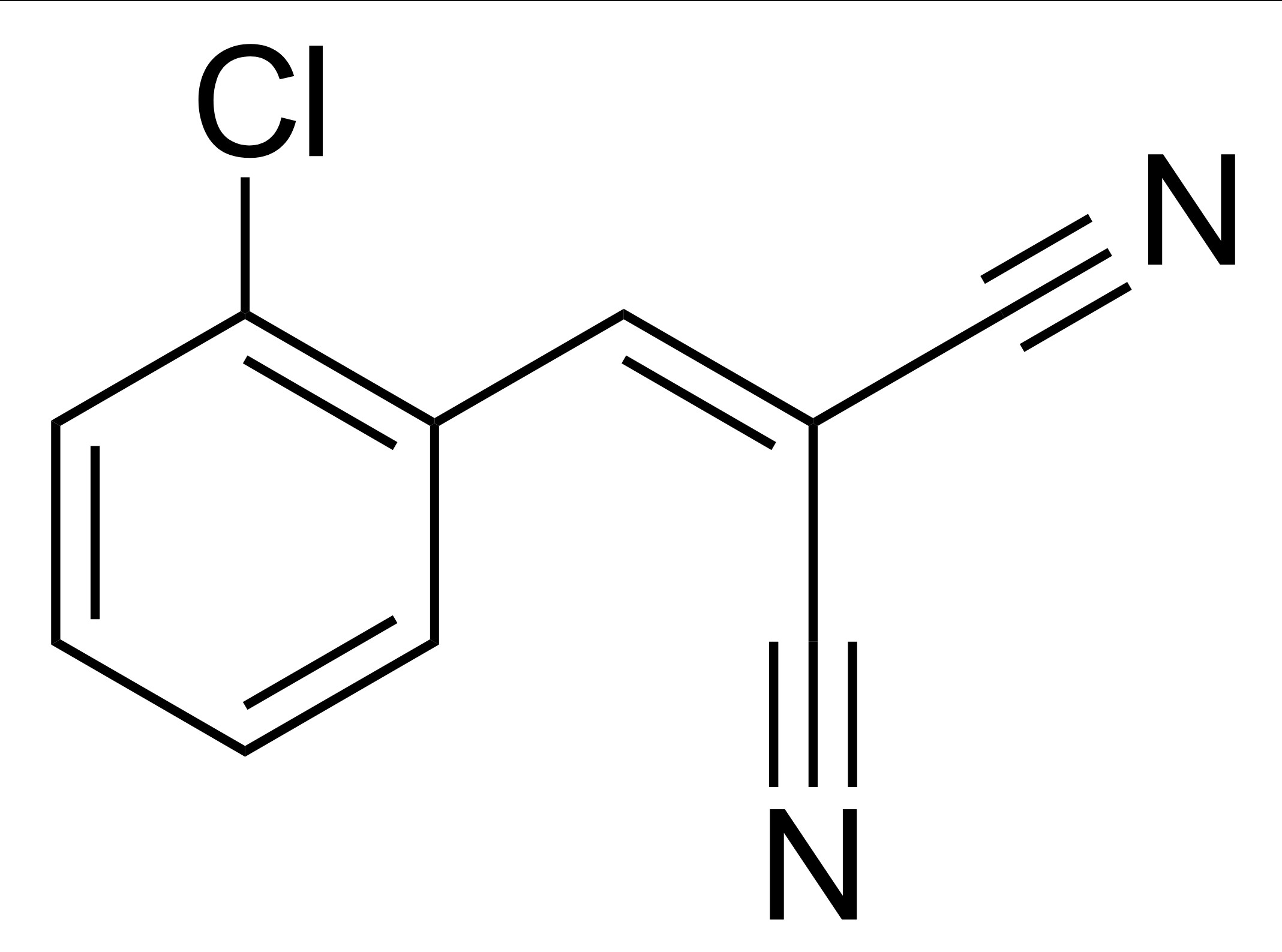
The chemical structure of tear gas irritant 2-chlorobenzalmalononitrile
Via Wikipedia
People exposed to tear gas were at risk for at least three months post-exposure for chest tightness, difficulty breathing, and chronic bronchitis. Severe exposure can cause edema and respiratory arrest leading to death as well. The CDC reports that tear gas can also kill due to severe burns in the respiratory tract. Other effects include corneal abrasion in the eye, glaucoma, and nerve damage. Deaths from deploying tear gas have also been reported after their use in prisons. A four month-old infant developed pneumonitis after two to three hours of exposure to tear gas, when police fired canisters into a home trying to arrest an adult there.
Tear gas was banned in warfare under the 1993 Geneva Protocol. Police continue to use it domestically, in order to avoid using more lethal tactics for crowd control.
Denisovans left their DNA traces in humans, but their fossils remain elusive
New study digs into ancestries of people in Island Southeast Asia
Extensive fossil and DNA evidence from around the world has shown that extinct human species occasionally interbred when they encountered one another. Modern humans interbred with at least two other species — Neanderthals and Denisovans — which have left traces in our DNA to this day.
In contrast to the well-studied Neanderthals, the Denisovans are a poorly understood species known from only a handful of 50,000 to 160,000 year old fossils from Siberia and Tibet. But genetic studies have shown that Denisovans interbred with modern humans in Island Southeast Asia (ISEA), thousands of miles from where all known fossils were found.
There are plenty of fossils in ISEA, belonging to three distinct species: the well-traveled Homo erectus, and two endemic "super-archaic" species. The last of these species have a deep history in the region but disappeared from the fossil record around 50,000 years ago. This combination of super-archaic fossils and Denisovan DNA has complicated our understanding of the history of this region. To disentangle the genetic relationships among human species in ISEA, an international team of scientists analyzed DNA of over 400 people from across the world, searching for segments that corresponded to both super-archaic and Denisovan DNA.
Surprisingly, they found no evidence that any of these super-archaic species interbred with modern humans, despite the wealth of fossils from the area. On the contrary, people with ancestry from ISEA, Papua New Guinea, and Australia had the largest amounts of Denisovan ancestry of all populations studied.
Finding genetic traces of Denisovans in an area of the world where they have yet to be found shows how little we know about the history of this region. While Denisovans appear anatomically distinct from the three super-archaic species of ISEA, it’s possible that they’ve been hiding in plain sight all along. There may also be Denisovan fossils still hidden across ISEA, waiting to be found. Either possibility will have major impacts on how we understand our own evolutionary history.
The muon g-2 experiment might mean the Standard Model of physics is incomplete, but that's just the beginning
Muons, elementary particles similar to electrons, appear to behave like magnets, but the Fermilab experiment is still not confirmation
Reidar Hahn via DoE
Twenty years ago, an experiment at Brookhaven National Lab produced some puzzling results that might point to new physics beyond our current understanding.
Just last week, Fermilab unveiled the product of a decade-long quest to verify that original Brookhaven result: they can now say with even more certainty that there is indeed a discrepancy between the measurement and our theoretical predictions based on current models of physics.
The experiment was a measurement of the so-called "muon g-2 factor." The muon — a subatomic particle which is, essentially, an electron, but 200 times heavier — has a magnetic moment, meaning this tiny particle can be thought of as a little bar magnet. Our current working theory of particle physics, called the Standard Model, predicts how strong this bar magnet should be. But that 20-year-old experiment measured a strength of the magnetic moment that wasn't consistent with the Standard Model theory prediction. Fermilab repeated the experiment with even more care and precision, by looking at how fast the muons' little bar magnets move like a spinning top when placed in a magnetic field. They were able to confirm that there is a 4.2-standard deviation discrepancy between their measurement and the current best theory predictions, meaning there is only a one-in-40,000 chance that the discrepancy is a statistical fluke.
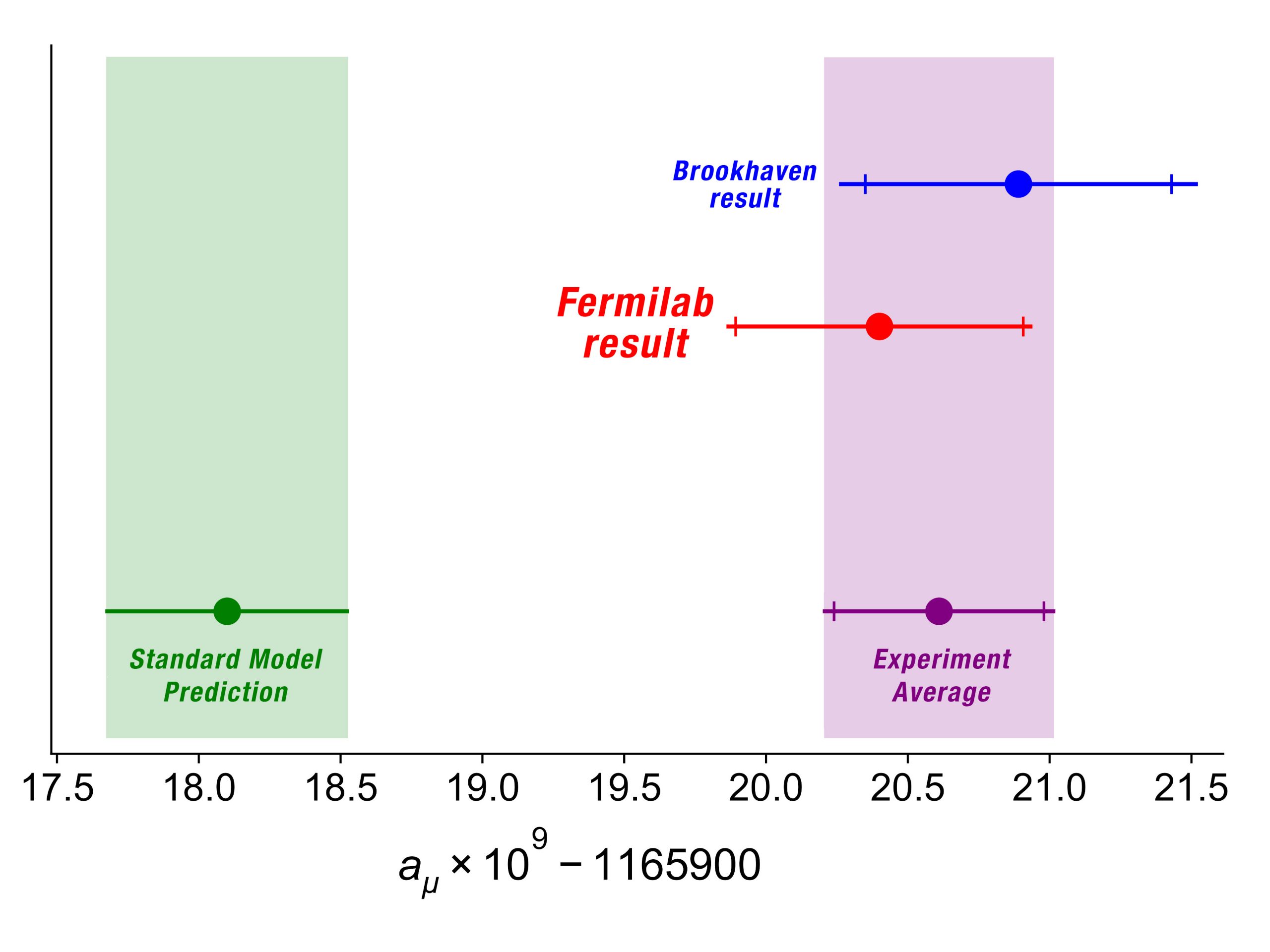
The Fermilab result from the Muon g-2 experiment at Fermilab aligns well with the previous Brookhaven result
Ryan Postel, Fermilab/Muon g-2 collaboration
This is exciting, because it could be a hint towards a new, more complete theory of physics that might answer many questions that the Standard Model hasn't been able to. But then again, there could just be an issue with the way that the muon's magnetic moment is being calculated, not a problem with the physics itself. Calculating the "g-2" (pronounced "g minus two") is extremely complicated, and not every method of calculation produces the same result. Last week, at the same time the new experimental value of the muon g-2 was announced, a theoretical value using a new method was also announced — and this new theoretical value is actually much closer to the measured value thank previous calculations have been.
So while this is a very important step in particle physics, it's by no means the end of the story. The theorists will be hard at work trying to figure out why different calculations are giving different results, and the experimentalists will be hard at work with the analysis of even more data runs, reducing their uncertainty further. We can expect more exciting announcements from Fermilab in the years to come.
Researchers identify a bacterium that enables its host to breathe nitrate instead of oxygen
Meet Candidatus Azoamicus ciliaticola, discovered in a Swiss lake
Collage from Wikimedia Commons, CC BY-SA 4.0
Most eukaryotes generate energy through breathing oxygen. This happens in mitochondria, specialized organelles likely acquired by the ancestor of eukaryotes when it engulfed a free-living prokaryote. Over time, this prokaryote became an obligate endosymbiont, meaning that neither the prokaryote nor its eukaryotic host could survive without the other. Then with more time it became an organelle, by keeping only the genes needed for oxygen respiration and losing genes for independent living.
But not all eukaryotes breathe oxygen. In a paper published in Nature, researchers investigating a lake in Switzerland found many eukaryotic ciliates (single-celled eukaryotic organisms covered with tiny hairs they use to move) which swam away from oxygen, indicating that they used something else for energy.
When the researchers stained the ciliates with a DNA-binding dye, they found multiple pockets of DNA within each ciliate outside of its nucleus. Using a different fluorescent dye, they found that these pockets contained only bacterial DNA. The researchers extracted and sequenced the DNA, and obtained a small circular genome not belonging to any known bacterium. They named this novel organism ‘Candidatus Azoamicus ciliaticola’.
Why was this bacterium inside these ciliates? The researchers compared its genome to other bacteria to answer this question. Like many other endosymbionts, the bacterium's genome was very small and lacked the genes needed for independent living. In particular, this genome contained a high proportion of genes for energy production, similar to mitochondrial genomes. However, it lacked genes for oxygen respiration. It instead possessed the genes needed for nitrate respiration. These clues led the researchers to conclude that ‘Candidatus Azoamicus ciliaticola’ was an obligate endosymbiont that enabled its ciliate host to breathe nitrate — not oxygen — for energy. This adaptation allows the ciliate to live in waters low in oxygen but high in nitrate.
While many free-living prokaryotes can respire nitrate, this is the first instance of this metabolism in a prokaryotic endosymbiont. Perhaps other eukaryotes out there have acquired new metabolisms with the help of endosymbionts. These interkingdom partnerships may allow eukaryotes to colonize environments formerly assumed to be the domain of only bacteria and archaea.
The less you know about a piece of art, the more impact it might make on you emotionally
This is your brain on poetry
Photo by Maria Lupan on Unsplash
Remember in high school, when you had to read the entire Divine Comedy — and said to yourself, “I’ll never major in literature?"
Unsurprisingly, brain recordings from experts in a particular discipline — be it literature, painting or music — show that experts value the types of art that interest them more positively than do people with little background on it.
But, just because you didn’t major in literature doesn’t mean that you can’t be moved by a poem. In a recent study conducted in Italy and published in the journal Brain Sciences, non-literature students showed higher emotional reaction to excerpts from the Divine Comedy than did literature students, despite the fact that the literature students were able to appreciate and recall it better. The researchers attribute this to 'emotional attenuation' in the literature students.
So if you are reading a poem and wishing you knew more, think again: it might be more emotionally impactful on you, the less you know about it.
People with immune systems primed to fend off bacteria are more susceptible to a common virus
This new research could lead to greater understanding of how the flu and coronaviruses affect us
Photo by Kelly Sikkema on Unsplash
Every winter coughs and sneezes run rampant through the population, but some people find themselves sick with the common cold time and again whereas others do not. Although immune memory generated from previous infections can partially explain why some people might be protected, it is not sufficient to explain the whole story.
Researchers at Imperial College London trying to better understand what makes us susceptible to the common cold, looked at the state of the airways in healthy volunteers before exposing them to Respiratory Syncytial Virus (RSV). RSV is one of a number of viruses which can cause common cold symptoms in healthy adults, but it can prove fatal in infants and the elderly.
In a paper published in Science, these researchers showed that people with bacteria-fighting cells, called neutrophils, in their airways before viral exposure were more likely to become infected with RSV. In this case, by being primed to tackle bacterial infections, their immune systems were less prepared to fight off a virus.
This finding could be used to identify people who might be more likely to become infected with RSV as well as further our understanding of how viruses affect us. These findings may prove relevant for other viruses such as influenza and coronaviruses.
Such a discovery would not have been possible without a challenge study in which study volunteers are safely exposed to the virus. This allows researchers to capture information before, during and after viral exposure, something which researchers at Imperial College London now hope to do with Sars-CoV-2, the virus responsible for the COVID-19 pandemic.
People of the Tiahuanaco civilization engineered their own rocks to build temples and monuments
The discovery illustrates the great ingenuity of ancient construction workers
Brattarb via Wikimedia Commons.
Tiahuanaco is a small village south-east of Lake Titicaca, is a world-famous archeological site with 1,400-year-old monuments and ceremonial buildings. The precision and detail of these sculptures called the attention of scientists who doubted that they could have been created with the simple tool technology known at the time.
A new pre-print, a scientific study that is completed but have not yet been peer-reviewed by other scientists, claims to solve the mystery: the ‘H shaped’ blocks of one of the most iconic temples ‘Pumapunku’ is not made of rock (as it was always thought), they’re made of sand!
Since archaeologists found the ruins of Tiahuanaco, they tried to find their source of materials, as almost all of the ancient city was made of rock blocks. In 1892 researchers discovered that these rocks (called andesite) were collected from an outcrop located at the foot of a volcano named “Cerro Khapia.”
A geochemical analysis revealed that the composition of the H shaped blocks from Pumapunku coincided with the andesitic sand but that also included organo-mineral binders, bat droppings, and other ingredients used to produce andesite geopolymer blocks.
The discovery of Tiahuanaco’s material supply route revealed the great ingenuity of ancient construction workers who created incredibly resistant blocks by using what they could find around them in ways archeologists had not anticipated.
New earthquake-detecting app may give people more time to evacuate
ShakeAlert was designed by researchers at the University of Oregon
Z22 on Wikimedia Commons.
The 10 year anniversary of the terrifying Tohoku earthquake and tsunami wrecked Japan passed by last month. Now, the other side of the Pacific Rim may have developed a new technology to better detect these devastating forces of nature.
ShakeAlert, designed by researchers at the University of Oregon and led by UO geophysicist Doug Toomey, uses the data from over 400 different seismic detectors (the ‘rumbles’ that proceed an earthquake) in the Pacific Northwest to be sent to smart phones and other wireless devices in areas where an earthquake might strike, including states such as California, Oregon, and Washington. Subsequently, by giving residents more time to prepare for an earthquake and even tsunami, more lives may be saved, similar to the impact that tornado warnings have had in preventing deaths from wind storms. Hopefully, this app can also expand to other countries and places vulnerable to earthquakes, such as Chile, New Zealand, and Japan, where notices of seismic activity can be sent quickly to people, allowing them to keep themselves and their loved ones safe during an otherwise cataclysmic event.
Uranus emits extra x-rays, and scientists don't know why
They could be just reflections, or Uranus could have its own version of the Northern Lights
Chandra image gallery. X-ray: NASA/CXO/University College London/W. Dunn et al; Optical: W.M. Keck Observatory.
Scientists have recently discovered x-rays coming from the planet Uranus. Using data from the Chandra X-Ray Telescope, scientists have observed x-rays on Uranus in images from both 2002 and 2017. (You might be thinking, “2002, how is that new?!” Sometimes, like in this case, astronomers will record a lot of data and not actually finish analyzing it until years later).
They combined this x-ray information (shown in pink) with optical pictures of Uranus (blue), resulting in the image you see here.

Combined optical and X-ray image of Uranus
Chandra image gallery. X-ray: NASA/CXO/University College London/W. Dunn et al; Optical: W.M. Keck Observatory
High energy x-rays from a planet might sound shocking, but we’ve actually seen x-rays coming from most planets in the solar system. The Sun emits x-rays, and planets reflect some of that light back into space. The interesting thing with Uranus is that it seems to show more x-ray than you’d expect from just reflected sunlight. So, how is Uranus producing extra x-rays? Maybe it simply reflects more x-ray light than the other planets, or maybe it has charged particles hitting its rings, like Saturn. Another explanation could be aurorae — like the Aurora Borealis on Earth, other planets emit light when charged particles (like electrons) travel along the lines of their magnetic fields.
Either way, we’ll need more observations to know for certain what’s going on with Uranus. We know quite a bit about our solar system, but the two ice giants (Uranus and Neptune) are woefully unexplored. The only mission to visit them was Voyager 2, back in the 1970s, and we haven’t been back since.
Scientists engineer a sustainable plastic made of tiny building blocks
The new plastic is just as strong as polyethylene and can be 3D printed into objects
Fran Jacquier on Unsplash.
Plastics are incredible because they are so versatile. Plastic waste, however, is problematic because it is so persistent. Right now, the life of a typical piece of plastic follows a linear path: plastic is born from fossil-fuel building-blocks, molded into products, and dumped into landfills and oceans as waste.
A cyclic life would be more sustainable, where plastic is born from plant-based building-blocks, molded into products, broken back down into building-blocks, reborn into plastic, and so on. This “closed-loop” recycling is challenging because the plastic must be durable enough for product use but breakable under the right conditions.
To obtain durable yet breakable material, researchers made new plastic that imitates the world’s most common plastic, polyethylene, but with a slightly different chemical make-up. Polyethylene is durable because it is made of carbon and hydrogen atoms, which form strong bonds with each other. Long chains of connected atoms in polyethylene also arrange themselves into an organized 3D structure, further increasing strength.
The new plastic still has long segments with carbon and hydrogen atoms, like polyethylene, but with small amounts of oxygen atoms, which serve as breaking points. To obtain a strong plastic despite these breaking points, the plastic needed to be made of extremely long chains of connected atoms. Researchers overcame this challenge by using a more reactive combination of molecular building-blocks to grow longer chains.
The new plastic is just as strong as polyethylene and was 3D printed into a protective smartphone cover and a cup that could withstand boiling water. However, a combination of heat and alcohol completely broke the new plastic down into its molecular building-blocks – even in mixtures with other plastics and dyes that are present in real-world waste streams. The building-blocks were then re-used to make the plastic again in a “closed-loop” lifecycle.
Plastics with “closed-loop” lifecycles could dramatically reduce the resources needed for products that we use every day, like smartphone covers or cups. While more research is needed to understand how to produce and recycle new plastics on large scales, this initial example is a promising step towards a sustainable future.
Space travel is now open to the public
SpaceX's Inspiration4 has picked two new astronauts and will be launching September 2021
Via press release
SpaceX’s Inspiration4 is the first all civilian space mission, breaking from a long tradition of NASA leadership and military-trained pilots. Funded by a billionaire CEO (Jared Isaacman, also the mission commander), the mission team just announced its final two crew members to join Isaacman and physician assistant and cancer survivor Hayley Arceneaux on the journey.
Dr. Sian Proctor, a geoscientist and educator, won her spot through an entrepreneurship competition for “demonstrating innovation and ingenuity” using Isaacman’s Shift4Shop platform. She is extremely well-qualified to be an astronaut, too — she completed multiple trips on Earth as an “analog astronaut” and has been a finalist in NASA’s Astronaut Program. Chris Sembroski, an engineer, Air Force veteran, and all-around space enthusiast, won his seat through a raffle where proceeds went to St. Jude’s.
Launch is planned for September 2021, and these new astronauts will orbit Earth every 90 minutes for multiple days in SpaceX’s Dragon crew capsule before returning home. This is by no means the first historic feat SpaceX has taken on — they were the first private company to launch a rocket successfully and the first to send astronauts to the International Space Station. They’ve also created the first reusable rocket, the Falcon 9, and currently have the largest satellite constellation in the world (although that one might not be such a good thing).
Despite their illustrious track record, they’re not the first company to get into space tourism — although they are the first to successfully make it happen without the government’s help. Billionaires have been buying tickets through Space Adventures (hopping aboard Soyuz spacecraft) for years, Jeff Bezos (of Amazon infamy) owns Blue Origin, and Sir Richard Branson has Virgin Galactic. Even more private space companies have appeared in recent years, showing us that we truly are on the verge of a new era of space exploration (for better or for worse), with SpaceX leading the way.
Think twice before you ice after an injury
Applying ice to a sprained ankle or wrist decreases blood flow to the area for longer than previously thought
Photo by Yogendra Singh on Unsplash
It is common practice to apply an ice pack to a sprained ankle or a sore muscle, and many professional athletes have been reported to use cryotherapy to aid with recovery. However, the benefits of the well-known RICE protocol (rest, ice, compression, and elevation) for injuries and sore muscles have been thoroughly debunked, including by the doctor that originally coined the term four decades ago. While icing an injury does effectively relieve pain, it also constricts blood vessels and reduces blood flow to the cold area. Even though the injury feels better, this impairs the body’s ability to heal, extending the recovery process.
But what happens after the ice is removed? In a recent study, scientists hypothesized that once the area warmed up, there would be a large temporary increase in blood flow, aiding in the healing process. This “rebound” phenomenon has been observed after things like removing a tourniquet or unclamping an artery during surgery, but hadn’t been studied for restrictions due to cold temperatures.
The researchers found that using ice, compression, and elevation therapy on a muscle immediately after exercise led to significantly reduced blood flow as expected, but instead of bouncing back immediately after treatment, the blood flow remained low for an extended period of time. While we already knew that ice impairs muscle recovery even though it’s great for reducing pain, now we can add that the negative effects last longer than previously hypothesized, suggesting that injured athletes should think twice before using ice as pain relief.
Hurricanes cause more health problems than you might think
New research into when and why people go to hospitals after hurricanes will help these facilities better prepare for future disasters
Daniel H. Farrell, U.S. Air National Guard on Flickr
Knowing how to prepare for a hurricane isn’t easy, especially if you are running a hospital. In the United States, hospital facilities are required by law to plan for disasters, including outlining emergency leadership and staffing, triage protocols, and how to perform a full-scale evacuation, as several Gulf Coast hospitals did during Hurricane Laura last fall. Making the wrong decision can cost dollars and lives, and there are always lessons to be learned.
A recent study published in Nature Communications may have one of those lessons. By cross-referencing 70 million Medicare hospitalizations with 16 years of local wind measures, researchers identified patterns of when older Americans tend to go to hospitals after storms and why. Unsurprisingly, hospital visits for injuries jumped post-storm. But so did several seemingly unrelated maladies.
The day after hurricane force winds, for instance, hospitalization rates for respiratory issues — asthma, chronic obstructive pulmonary disease (COPD) — doubled on average and infectious disease visits spiked by roughly half. Visits for both remained higher than average throughout the week after hurricanes and tropical cyclones, as did hospitalizations for liver disease, delirium and dementia, and renal failure. Cancer visits dropped by 4 percent in the same time.
All told, an estimated 16,000 additional hospitalizations were associated with respiratory problems and 2200 with infectious disease. Reasons are varied but likely have to do with the loss of power needed for breathing equipment and exposure to contaminated flood water — more than 800 wastewater facilities reported spills after Hurricane Harvey in 2017.
The results will help improve hospital preparedness for hurricanes. They will also clue hospitals in to how to stock medical supplies in the days before a storm and assign staff in the days after, crucial information when these facilities are strained. At a time when climate change is fueling more frequent and powerful cyclones, the study is also a reminder of the detrimental effect global warming has on health, even in wealthy countries.
Scientists re-discover a long carbon chain molecule hiding in space
Cyanodecapentayne has been detected in the Taurus Molecular Cloud for the first time in over 20 years
ESA/Herschel/NASA/JPL-Caltech; R. Hurt (JPL-Caltech) (CC BY-SA 3.0 IGO)
Cyanopolyynes are a class of molecules that are out of this world. No, really — these molecules do not exist naturally on present-day Earth.
Cyanopolyynes are long chains of carbon atoms with a hydrogen atom on one end and a nitrogen atom on the other. Interstellar cyanopolyynes are interesting because they might help us better understand the carbon chemistry around stars and how larger carbon molecules, such as those that make up interstellar dust grains and soot, are formed and destroyed during stellar evolution.
Observations with the Green Bank Telescope (GBT) in West Virginia have revealed another interstellar cyanopolyyne - cyanodecapentayne (pronounced sigh-ann-oh-deck-uh-pent-uh-ine), or HC11N - in the Taurus Molecular Cloud, about 430 light years from Earth. This discovery was reported in the February 2021 issue of Nature Astronomy, but the saga of detecting this molecule goes back to the late 1970s.
Predictions for HC11N’s observable chemical signature were reported in 1978. Throughout the 1980s and 1990s, there were multiple detections of HC11N around a cool star and in the Taurus Molecular Cloud, but more recent observations taken toward that same cloud came up empty. This suggested that the earlier reports were based on a mistake made in the assumptions of the molecule’s chemical signature. After more than 20 years, HC11N was knocked off the list of detected molecules, until this latest discovery.
Long carbon chains up to HC17N have been detected in the lab, so there might be even longer carbon chain molecules hiding in the clouds of interstellar space. The bigger question is: will we be able to detect them?
A scientific correction finds Venus's atmosphere probably does not contain phosphine gas
The initial discovery set off a flurry of excitement. The reality is something more mundane
Is there life on Venus? We once envisaged an alien world hidden beneath its yellow clouds. Through advances in astronomy, we now know that Venus is rather uninhabitable. It's scorching hot with a toxic sulfur dioxide clouds. The air pressure on the surface is literally crushing.
A study in September 2020 noticed an anomaly in the atmosphere of Venus. By pointing a telescope at the planet, researchers detected much more phosphine gas than expected in its atmosphere. This report set of a flurry of publicity and excitement, because the only way we know of to make this much gas involves microbial processes. The scientists were very careful in writing about their findings, conscious of the fact that extraordinary claims require extraordinary evidence, but it was widely hypothesized that either this was a newly discovered reaction in the atmosphere or a sign of microbes on Venus.
But in November of 2020, the authors contacted the journal to say that they found mistakes in the way that they processed some of their telescope data. They are in the process of correcting their article. In the meantime, other groups also delved into these findings. In January 2021, another group of researchers published their analysis, and they concluded that the gas was likely sulfur dioxide, not phosphine, high up in Venus's atmosphere.
So, scientists didn't find signs of alien life on Venus after all. Nonetheless, they managed to improve their techniques and calibrations for observing faraway planets. And — importantly — the scientific process of discovery, debate, and correction when necessary worked exactly as it should have.
By avoiding pumas, deer are drastically changing the vegetation around them
Animals' behaviors are shaped by fear of humans, and this, in turn, affects the plants they eat
Photo by Priscilla Du Preez on Unsplash
We humans modify our surroundings to fit our needs. We are capable of taking out any animal, from an inconvenient garden snail to a wolf that has a habit of killing livestock. It is no wonder that animals fear us.
But the effects of that fear do not stop with the animals themselves. Animals are adaptable creatures. They adapt to our schedules and behaviors by modifying their own. These changes in behaviour can trickle down. These ecological effects extend even to large carnivores like pumas.
Pumas live throughout the Americas, from Patagonia in the south all the way to the sub-arctic regions of Canada. But most people who live in those areas will go their whole lives never seeing one. Pumas know to avoid us — and their prey have noticed too.
New research published in Ecosphere has found that black-tailed deer in the Santa Cruz Mountains of California are spending more time in the forest areas closer to human habitation. And it is not due to tastier or better quality plants, nor because humans are providing them with food. Rather, it is because pumas are afraid of the nearby humans and so they don’t like going into those areas.
The deer are spending so much time in these forests that they are modifying the environment. The plants in these deer-frequented areas are becoming shrubbier as they are essentially being pruned by the deer. This, in turn, creates more food for deer, creating a win-win scenario for the deer. More research is needed to see how this affects the other wildlife in the area, like birds and insects, but the effect that humans are indirectly having on the landscape is clear.
Orangutans and bonobos at the San Diego Zoo have been vaccinated for COVID-19
The apes were given an experimental vaccine originally developed for dogs and cats
Photo by Simon Infanger on Unsplash
Just over three months ago, in December 2020, research found that monkeys and apes from Africa and Asia are susceptible to the SARS-CoV-2 virus. At the time, there was particular concern for endangered gorillas because the disease could have a detrimental effect on their small population size, and for captive chimpanzees living in sanctuaries who are in close contact with humans.
Then in January, it was reported that a troop of eight captive gorillas living at the San Diego Zoo tested positive for COVID-19. While the gorillas made a full recovery, this incident showed that gorillas can, in fact, contract and become sick from COVID-19.
On March 3, however, there was good news for our primate cousins: great apes living at the San Diego Zoo received experimental COVID-19 vaccines. Four orangutans and five bonobos were vaccinated, making these nine animals the only non-human great apes to be immunized in the world. There are three leftover doses, which will be given to the three gorillas living at the zoo who did not have COVID-19 in January.
This specific vaccine cannot be used for humans. It was developed by Zoetis for dogs and cats, among ongoing debate as to whether such a vaccine would be useful. While the vaccine used on the great apes in San Diego was not specifically designed for them, Zoetis provided an emergency supply of the vaccine for this specific use. The idea of using a vaccine for one animal to inoculate another is not new. In fact, the influenza vaccine developed for humans is given to great apes in zoos every year and researchers are working to develop cross-species vaccines to treat other diseases like malaria.
While there is still relatively little known about how COVID-19 affects the animal kingdom, scientists are also worried about whether the virus could gain foothold in an animal population, and then reemerge and infect humans again. While it remains unclear whether the vaccine could (or should) be used to inoculate wild populations, the immunization of captive populations is certainly a step towards a safer world for all animals.
Two mistakes by a pair of astronomers expanded our knowledge of dying stars
Wade and Hjellming were initially looking for red supergiants. They didn't find them, but that doesn't mean they failed
beate bachmann on Pixabay
Radio telescopes detect signals at a longer wavelength than optical telescopes, making them ideal for looking at gases but not light or stars. In 1970, a pair of astronomers named Wade and Hjellming decided to use their three precious weeks of observing time on a radio telescope to see if a certain type of star called a red supergiant was detectable. After two weeks of searching, the answer was a disappointing “no.” They moved onto the backup plan: using their last week to point the telescope at various other types of stars in the hopes they could find something.
And they did! They detected a strong signal when they pointed the telescope at a nova, or exploding star, and then shortly after, a second one. Building on those successes they completely changed their observing strategy to focus on all types of novae, ultimately leading to the discovery of a new class of radio source and providing a valuable complement to the existing optical data on novae.
There was one small catch — they had mis-typed a digit when filling out the punch card that feeds coordinates to the telescope, and the first thing they had detected was actually radio waves from a known radio source, not a nova. Same thing for the second detection: a calculation error meant that they had been pointing at the wrong patch of sky. The fact that they kept looking specifically at novae with the radio telescope was propelled by a few silly mistakes that just happened to lead them down the right path of inquiry. As Wade wrote: “When you can’t do [science] any other way, that’s how you have to do it!”
The mysterious cause of sea star wasting syndrome is a mystery no more
Sea stars suffer when microorganisms living on them suck up too much oxygen from the water
Photo by Linus Nylund on Unsplash
Sea stars, also known as starfish, have a reputation for being resilient animals that can regenerate lost limbs. However, in 2013, sea stars off of the Pacific Coast began rapidly dying in huge numbers, and no one knew why.
Similar sea star mass die-offs have been recorded for decades, but this event was one of the largest wildlife mass-mortality events ever recorded. Sick sea stars become covered in lesions, permanently losing their limbs and melting into blobs of decayed tissue. This illness, termed sea star wasting syndrome (SSWS), was widely thought to be caused by a viral infection. However, this could not be replicated in the lab and was ultimately disproven.
Now, in a new study published in Frontiers in Microbiology, researchers have found the mysterious illness was caused by microorganisms sucking up oxygen from the water around infected sea stars, essentially suffocating them.
The researchers had previously investigated and ruled out other factors, such as water temperature, as the cause for SSWS. However, when they examined the water immediately surrounding sick sea stars to the environments around healthy sea stars, they found that nutrient-loving bacteria living on the sea stars had used up all the oxygen that they need to breathe.
Although SSWS is caused by an ecological interaction rather than an infection, it can still be transmitted between sea stars. As dying sea stars decay, they generate organic matter that can promote bacterial growth on nearby sea stars in a dangerous feedback loop.
Sea stars play essential roles in many ecosystems and help maintain local biodiversity. Knowing how this disease develops can help researchers treat sick sea stars in the lab, helping to preserve delicate ecological relationships.
Scientists starved E. coli for 1200 days to learn about bacterial evolution
They saw two main lineages emerge and compete with each other — all in a single test tube
Photo by Girl with red hat on Unsplash
What do you get when you stick E. coli in a test tube and don't feed it for three years? According to a new report in mBio, bacterial evolution at its finest.
In this study, scientists grew a single tube of E. coli in the laboratory for 1,200 days (over three years!). They wanted to explore how bacteria adapt to nutrient-limited conditions for an extended period of time, as microbes often face periods of starvation and stress in the natural world.
The growth medium in the tube was never replenished. The only nutrients available were those the bacteria recycled from waste accumulating in the tube. This approach was different from other long-term E. coli evolution experiments (some of which have gone on for over 25 years), in which bacteria are transferred to fresh media everyday.
When bacteria divide, they sometimes acquire mutations in their DNA. If those mutations are beneficial to the organism, their frequency within the population increases. By analyzing these mutations and their frequency, we can learn how bacteria evolve and adapt to their surroundings.
In this study, the researchers sequenced DNA collected from the E.coli collected at regular intervals over the course of the experiment. Based on the type and frequency of mutations they found, the researchers identified two main genetic groups, or lineages, that had diverged from the parent cells. While cells within each lineage coexisted with each other in the tube, one generally outnumbered those from the other, with the dominant lineage changing from one time point to the next. This indicates that the lineage best equipped to handle life in the tube varied according to the challenges faced by the cells at any given time.
In addition to being the first to explore evolution of a single bacterial population under nutrient starvation conditions for this length of time, this study illustrates how complex and dynamic evolving bacterial communities can be. It also highlights specific mutations that may help bacteria thrive in diverse, often stressful, environments in the world beyond the test tube.





















