Lab Notes
Short stories and links shared by the scientists in our community
Forget the Large Hadron Collider: fundamental particles ride plasma waves into high-speed collisions
If built, EuPRAXIA will be the world's first high energy plasma-based accelerator
By Jared Tarbell on Flickr
What comes to mind when you hear the words particle accelerator?
Here's what I think, in the order of the thoughts that show up in my mind: giant metal ducts, Higgs boson, The Big Bang Theory and the CERN. I bet you got some of these as well (if not all of them), and here is why.
The Large Hadron Collider (LHC), at CERN in Geneva, is the very same particle accelerator where the Higgs boson was first observed. With a circumference of 27 kilometres, the LHC is the one of the largest machines in the world. It is even common knowledge as it has made headlines, and even been mentioned in The Big Bang Theory more than once.
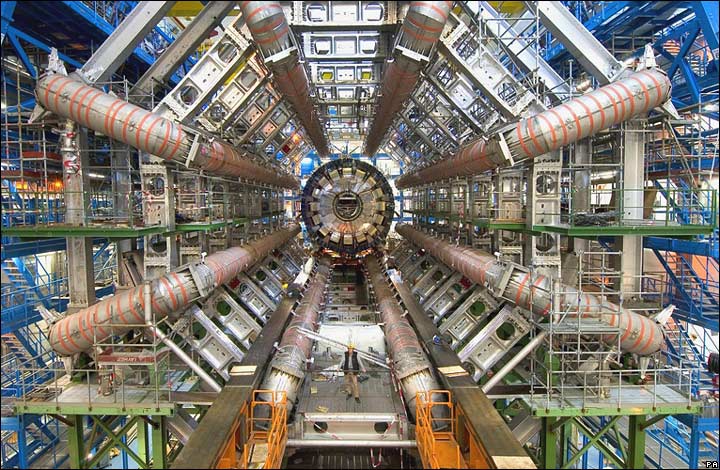
The Large Hadron Collider/ATLAS at CERN
However, did you know that approximately 30,000 particle accelerators exist today with many different functions? Not all of them are utilized for fundamental physics like the LHC — in fact only around 1% are. Instead, many of the smaller accelerators are used for cancer therapy, semiconductor manufacturing, or the production and identification of radioisotopes. In each case, the acceleration mechanism (which uses oscillating radio frequencies) requires large dimensions, coupled with high construction and maintenance costs, creating a limit on the use of such particle accelerators.
Instead, researchers in Europe are attempting to come up with a solution to bypass particle accelerators by using a plasma-based mechanism that should generate 1000 more acceleration potential per length.
Enter the European Plasma Research Accelerator with eXcellence, or EuPRAXIA for short.
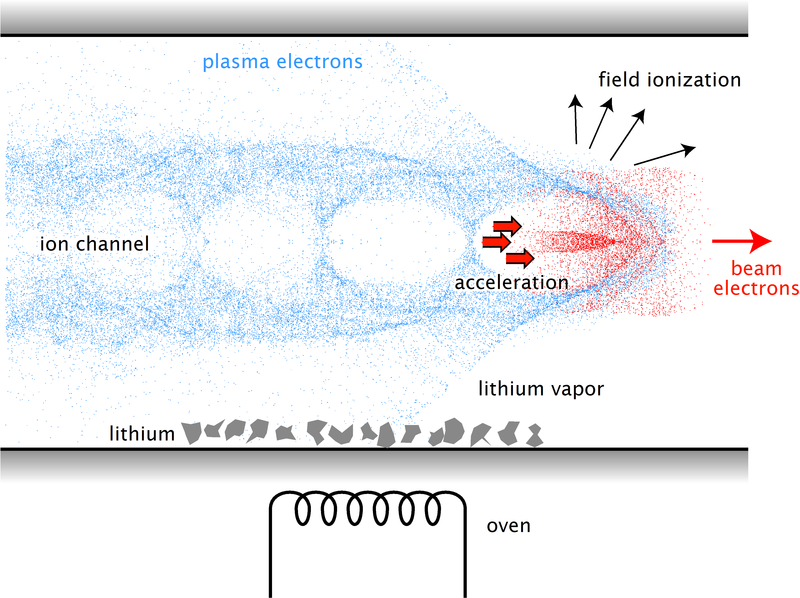
Illustration of the wake created by an electron beam in a plasma. This wake can be used to accelerate charged particles.
EuPRAXIA plans to use the electric-field gradients, which are created in a pulse's wake, to accelerate particles to high energy levels while travelling over very short distances. It will require smaller, more compact and cost-effective machines.
Currently, plasma-based particle accelerators are still undergoing a proof-of-concept phase at multiple institutions, including CERN. If built, EuPRAXIA will be the world's first high energy plasma-based accelerator. In a recent conceptual design report, presented by over 40 institutes who are collaborating on this project, the authors state:
“EuPRAXIA is designed to be the required stepping stone to possible future plasma-based facilities… [it] should be fully functioning in the next 8-10 years.”
The idea is to build a facility — that also external operators could use — where the many intriguing potentials of particle accelerators will be explored, such that, maybe one day, giant metal ducts will not be the first thing that comes to mind.
Why we shouldn't write off nuclear power
Despite radiation fears, changing to nuclear power could prevent millions of air pollution-associated deaths
Photo by Thomas Millot on Unsplash
After the "scare" near a Canadian nuclear power plant on January 12 which accidently sent a warning text to residents, many people are wondering about nuclear power. Is it safe?
In fact, nuclear power is one of the safest forms of power in terms of health burdens like air pollution. Even when you look at the larger picture of "death rate," nuclear energy is the third safest technology — behind only hydroelectricity and wind. In fact, in one study, it was estimated that the use of nuclear power has prevented about 1.84 million air pollution-related deaths. It has also been estimated, by the same study, that if nuclear power replaces fossil fuels, an additional 420,000 - 7.04 million future air pollution deaths could be prevented.

Photo by Johannes Plenio on Unsplash
And that's just the tip of the iceberg. Nuclear power is one of the most efficient energy systems we have access to — while natural gas or coal power plants operate at full power about 50% of the time, nuclear power plants operate at full power 92.3% of the time.
Now, many people are rightfully concerned about radiation (radioactivity is nothing to joke about). But, believe it or not, nuclear power plants release LESS radiation into the environment than coal power plants. That's right! In fact, the waste by-product produced by one coal power plant (known as fly ash), carries 100 times more radioactivity into the environment than the shielded waste produced by a comparable nuclear power plant.
Now, of course, nuclear power isn't without its drawbacks — the problem of what to do with spent fuel is one of them, and past nuclear disasters may weigh heavily on people's minds. But to write off nuclear power as inherently unsafe or not worth pursuing, overlooks the many advantages it has when compared to coal burning, which has serious consequences in terms of air and water pollution as well as greenhouse gas emissions.
Fish use "blood-doping" to survive in icy water
To keep their blood from becoming too thick, cold-water fish release blood cells from the spleen only when needed
NOAA Photo Library via Flickr
Being a fish in the Antarctic is hard. The bald notothen, a fish that hunts for prey under the ice in Antarctica, has come up with all kinds of tricks to be able to survive in the freezing waters. Like many fish that live in cold waters, the bald notothen has a kind of antifreeze protein in its blood, which prevents it from freezing. However, this strategy comes at a cost. The antifreeze in combination with the cold temperature causes the blood to become viscous.
“Due to this increased blood viscosity, the heart must work much harder,” explains Jeroen Brijs, lead author of a new study on bald notothen. In the long term, this could cause significant problems for the fish, but they’ve come up with a solution: their spleens are able to take up red blood cells. This reduces the number of these cells in the blood, so the blood becomes less viscous.

Photo by Osman Rana on Unsplash
But they need to use this technique carefully, because red blood cells are needed to transport oxygen, which the fish need to be active. Without the red blood cells, they are effectively anemic and unable to hunt or feed. The fish have solved this problem by using a dynamic strategy: they release the red blood cells when they need to be active, and take them up in times of rest. This tactic has made them extremely successful in the cold waters.
They are not the only fish that use this technique. “Although the blood boosting strategy of bald notothens is not unique within the notothenioids, the magnitude of the strategy in bald notothens is by far the greatest,” says Brijs. What the bald notothen does is effectively the same as blood doping in athletes, but the fish are much better at it. Athletes can only increase their oxygen carrying capacity by about 5-25 percent. The bald notothens can increase their ability to carry oxygen by as much as 207 percent.
In order to be successful in extreme environments, fish have evolved many strategies to survive and thrive. For many fish, blood doping might be the key to living in the cold seas.
Don't believe the conspiracy theories you hear about coronavirus and HIV
Especially if you work for the New York Times
By Mimi Thian on Unsplash
The new coronavirus is scary, and lots of information is flying around in the pandemic. But please, don't believe everything you read, even if it came from scientists. Before panicking about news, wait for real, actual confirmation (and peer review) from scientists outside of a Twitter scrum.
Case in point, today a group of scientists based in India took the genome sequences of 2019-nCoV, the novel coronavirus, did some armchair analysis (literally, all the work they did could be done from a desk in about an hour) which they shared in a pre-print (i.e. this hasn't gone through peer-review by other scientists yet), and found that it had similarities to the genome of...HIV.
Update, February 2nd: The paper has been withdrawn, following widespread criticism, like the kind below.
If you're a soup-brained Twitter user like me, you might have seen this sensational, zombie movie-ass tweet about the pre-print by Anand Ranganathan, a molecular biologist and author with 200k+ followers:
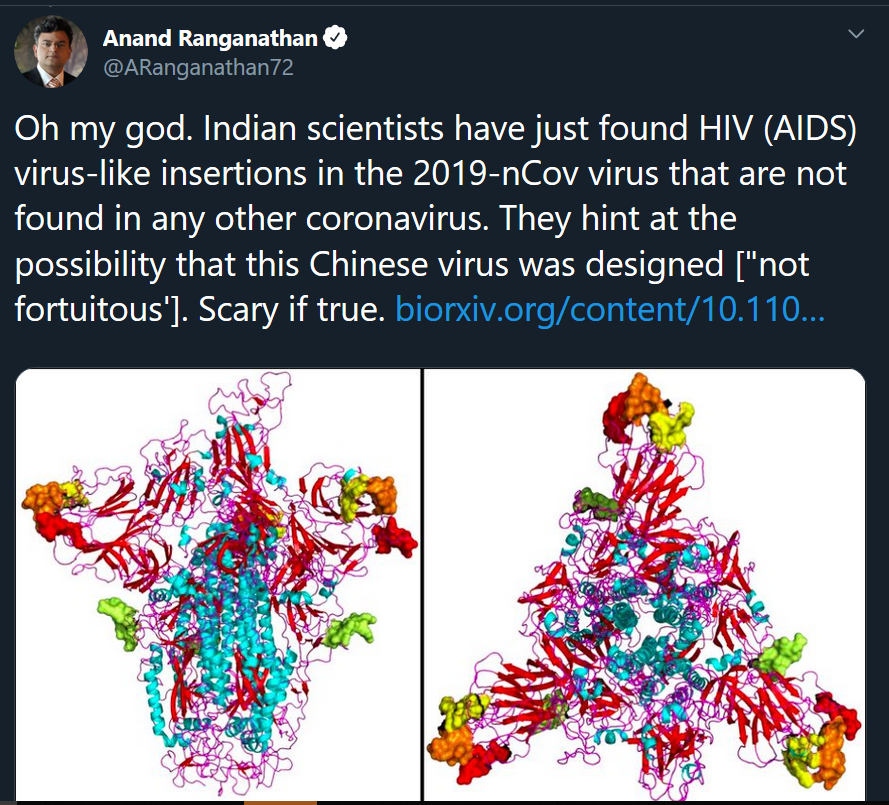
No, I'm not linking to it.
This prompted a blizzard of apocalyptic world building in the mentions, including a boost from prominent not-a-biologist New York Times opinion columnist Ross Douthat, who has about 160k followers:
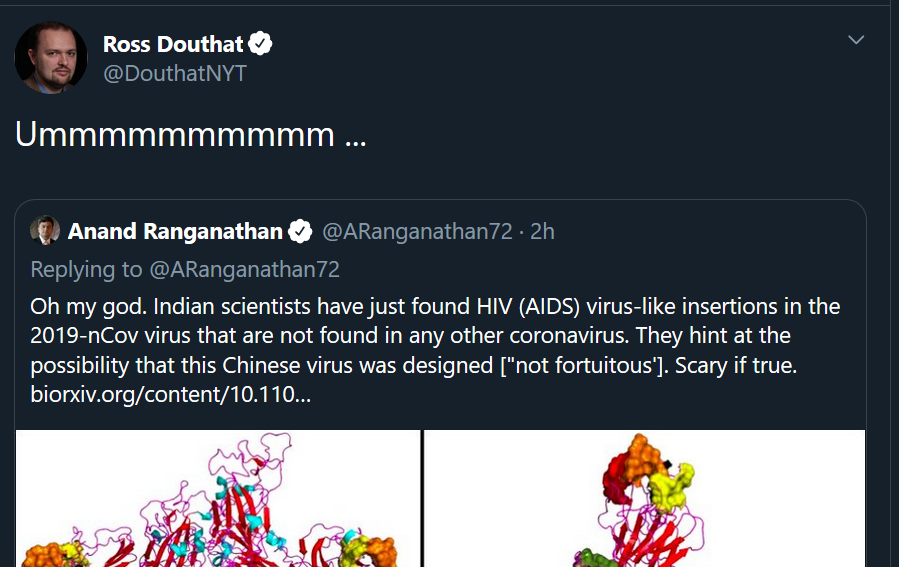
Thanks Ross, good input.
The gist of the story is this: the coronavirus (2019-nCoV) appears to have four additions to its genome that similar viruses, like SARS and MERS, are lacking. These additions are either identical or extremely similar to parts of the HIV genome. The authors go on to speculate that this may be why the coronavirus is spreading so quickly, and also that something sinister may be afoot:
"This uncanny similarity of novel inserts in [Wuhan coronavirus] to [HIV] is unlikely to be fortuitous."
They don't elaborate on what that means, but the Twitter interpretation is that someone engineered coronavirus with parts of HIV, to make it...I don't know. Infect better? It's unclear.
I double checked whether it was true that HIV and coronavirus have similarities. I took the four additional sequences (the "novel inserts") they found in the coronavirus genome and I looked to see if, as they said, it was similar to HIV. The truth is while these sequences do pop up in HIV, they also pop up in tons of other viruses. There's no reason to immediately suspect a nefarious fusion of HIV and coronavirus.
Sequence one out of four is indeed found in both coronavirus and HIV. The same sequence also appears in a virus that infects Streptococcus (spherical bacteria), a rat virus, and an "acute bee paralysis virus."
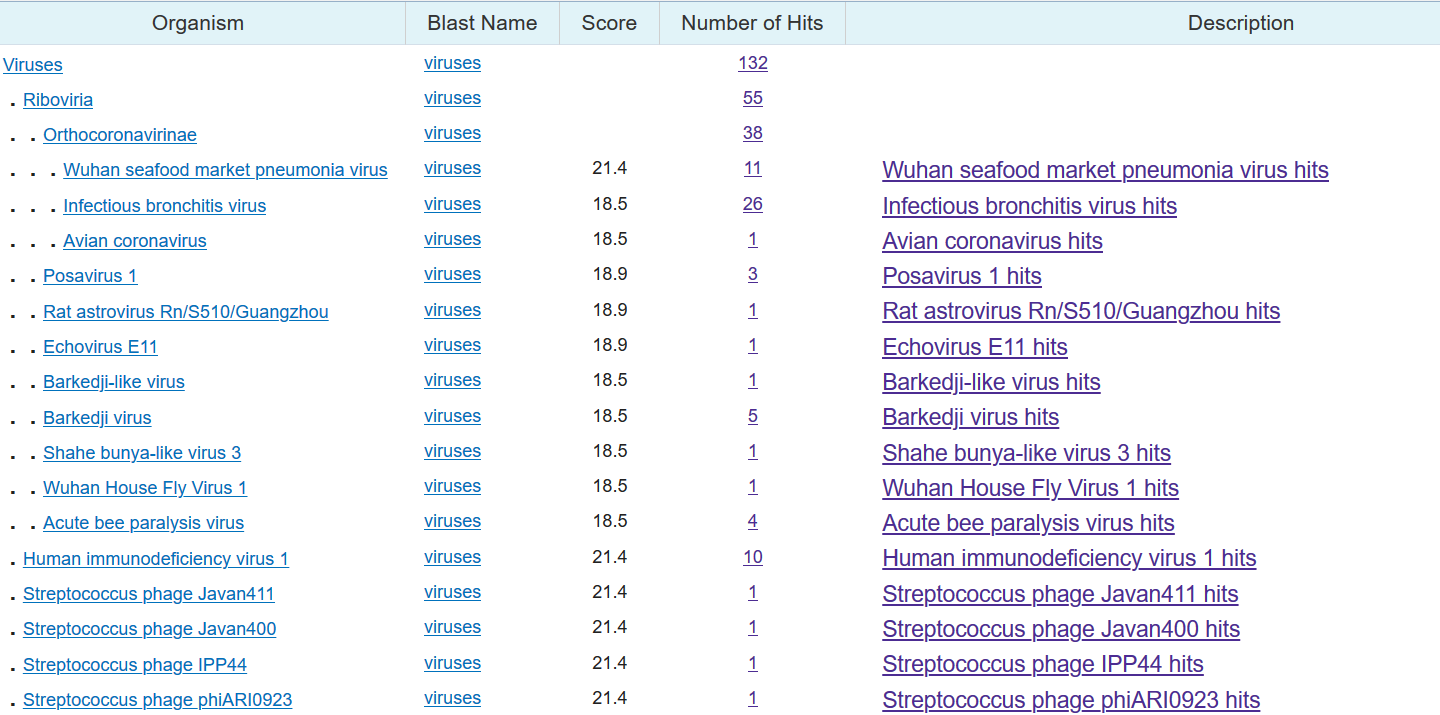
A list of viruses that contain 100% matches of sequence one from Wuhan coronavirus, which includes the coronavirus, HIV...and also a bunch of other viruses.
Dan Samorodnitsky
I did the same analysis on sequence two. Once again, HIV and the coronavirus appear together, but also a mouse herpes virus, a rat cytomegalovirus (the virus that causes mononucleosis, popularly known as mono or the 'kissing disease'), and a virus that infects sweet potatoes.
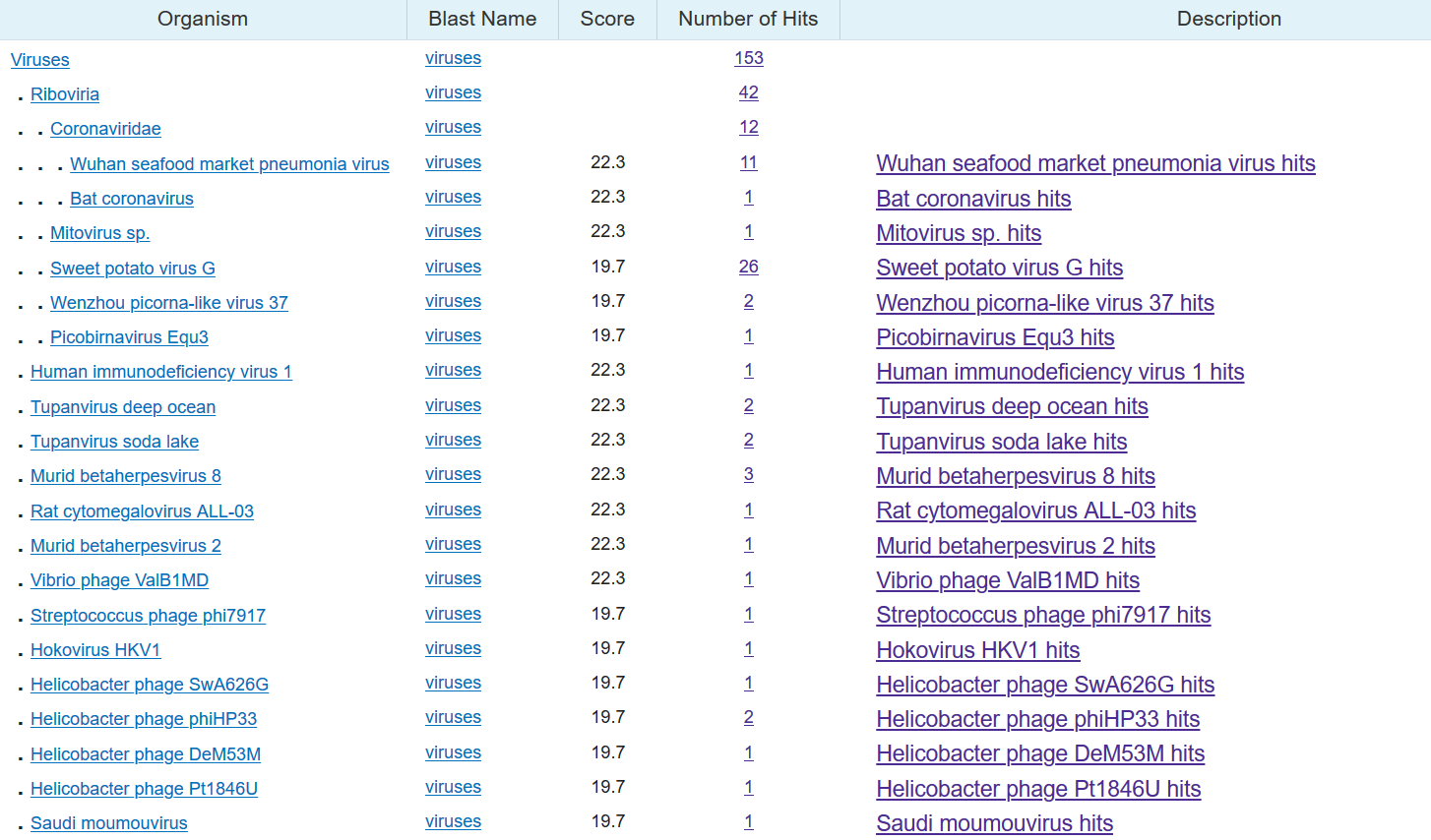
The stories for the other two sequences are the same. Those sequences appear in benign viruses like Peach-associated luteovirus (a plant virus), Bovine papillomavirus type 9 (the cow version of HPV), and Leptopilina boulardi Toti-like virus (a virus that infects wasps).
So the facts are that the new coronavirus may have some new additions to it, but they're common bits and pieces found in tons of viruses — not just HIV — and there's no proof that they even have anything to do with how the coronavirus behaves.
And there's definitely no proof that they were put there on purpose.
Algae are eating microplastics. From there, they can climb up the food chain to us
Freshwater microbes can absorb components of microplastics before getting eaten by other creatures
Photo by Jasmin Sessler on Unsplash
Microplastics have been making splashy headlines for years now and there's news: scientists used special carbon-containing polymers to track how microplastic moves through the aquatic biosphere. Plastics are very resistant to degradation in natural environments; knowing where they end up and how they're incorporated into food webs matters for human health. What starts out in algae can eventually end up in us.
To track how some microplastic moves through aquatic food webs, scientists in Finland and Austria turned to the molecular scale. They altered molecules of polyethelene (a common compound in plastics) to contain carbon-13, which they could use to trace those molecules later on. By measuring that tracer compound in the cell membranes of algae and zooplankton (aquatic microorganisms), they were able to see that carbon being incorporated into the organisms. As the zooplankton consumed the algae, the carbon went with them.

Photo by Ishan @seefromthesky on Unsplash
This is a step above how you might have seen plastic pollution before — a fish's stomach full of bits of bags and pieces of bottles. These microorganisms are actually repurposing the molecules in plastic, which increases the odds that other compounds in plastic could work their way up the food chain, too.
We can't have a future with quantum computers without quantum mechanics
Scientists are building physical platforms for memory, computation, and networking — all things we need for quantum computing to be possible
Kevin Ku on Unsplash
A future with quantum computers and quantum networks requires a diverse set of technical tools. Like the classical computers we use every day, we need physical platforms for memory, computation, and networking. Unlike classical computers, however, these platforms need to be quantum-mechanical (read: very small and very cold).
The verdict is still out on which physical platforms will work best for quantum computing, but scientists are making breakthroughs on the more promising ones. Just this year, a group in the UK coupled a single ion (charged atom) to a very strong light field for the first time. They achieved what atomic physicists call the “strong coupling regime,” where the ion is more likely to interact with the light field than to lose energy and information to the rest of its environment.
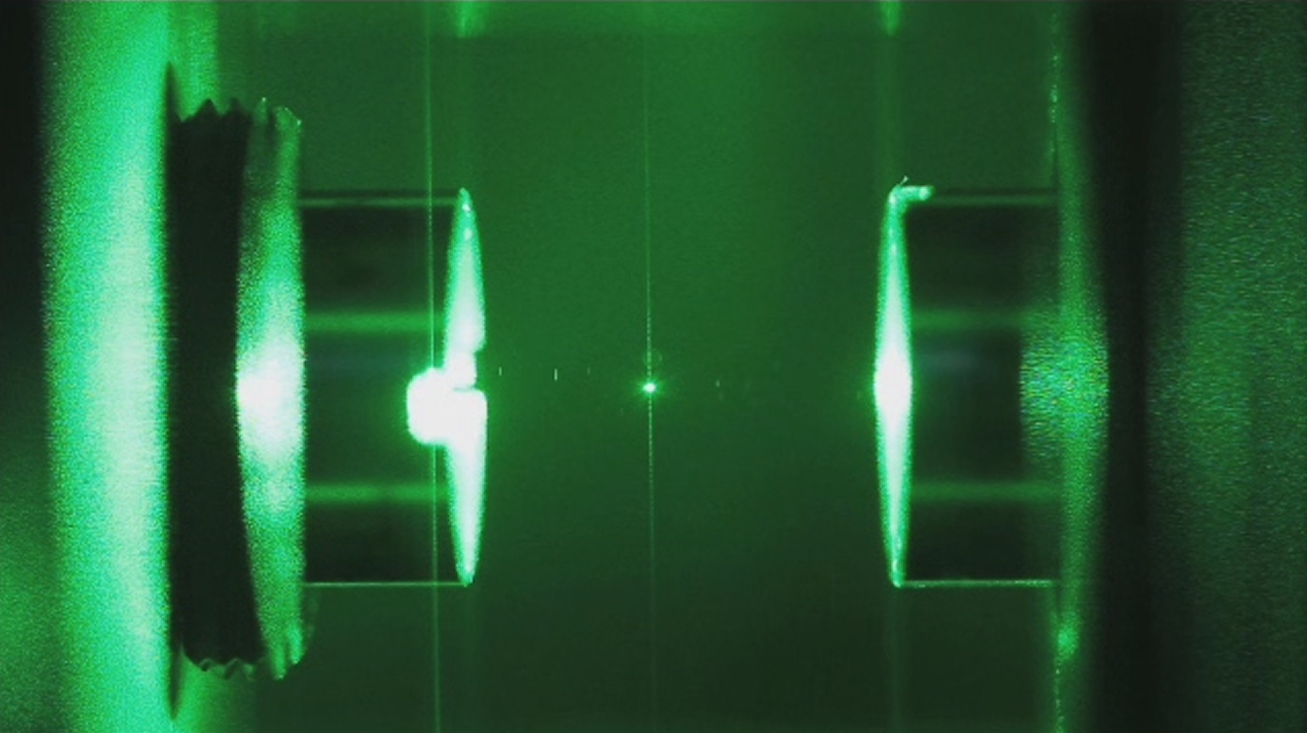
Here's a glass nanoparticle suspended in an optical cavity to help you picture what scale these scientists are working at.
By James Millen (UCL Physics & Astronomy at Wikimedia Commons
These physicists achieved the strong coupling regime by putting the trapped ion in an optical cavity: two mirrors facing each other that trap light for a number of round-trip reflections. The smaller the volume between these mirrors, the stronger the coupling between the light and the ion. By using optical fibers as mirrors, they were able to make this volume very small.
Light-ion coupling is exactly the sort of effect that will allow scientists to create quantum networks. To send quantum information between quantum computers, we can send photons with specific quantum states through optical fibers. However, optical fibers aren't perfect — photons lose information as they travel large distances.
By putting a strongly-coupled ion in an optical cavity at each of these nodes, we can transfer quantum states from an incoming photon to the ion and back to a new photon to send on its way. The advances in this paper increased the efficiency of an important step in this procedure (heralded entanglement) by a factor of 1,900 compared with previous iterations.
With a total efficiency of only 1.7%, however, there's still a lot of work to be done before the age of practical quantum networks.
My cat's coat is mostly white with dark tabby patches. What's going on?
A researcher uses a tweetorial to figure out the underlying genetics for their cat's coat
Photo by Paul Hanaoka on Unsplash
Have you ever wondered why your cat’s coat looks the way it does? Wonder no longer! Earlier this month, Emily Summerbell, a PhD student at Emory University, tweeted a ‘tweetorial’ on cat coat genetics. I used it to figure out the underlying genetics for my own cat’s coat.

Cat tax.
Adriaan Grijseels
This is my cat ‘Little Excuse’ (because when he’s sleeping on your lap, you’ve got an excuse to not get up).
Excuse has a short coat, which is controlled by the FGF5 gene. Short hair (L) is the dominant trait, meaning that if one out of two copies, or alleles, of the gene encodes for short hair, the cat will end up with short hair. Excuse is either homozygous, meaning he has two short hair alleles (LL), or heterozygous, meaning he has one short hair, and one long hair allele (Ll).
Though Excuse is for the majority white (we’ll get back to that later), he generally has dark-colored fur. This is encoded by the TYRP1 gene, for which the black allele is dominant. If he would have the allele for orange hair, this would override the black color, so the lack of any orange means that he has the recessive allele for the orange gene.

What type of coat does your cat have?
By Humberto Arellano on Unsplash
Back to the white fur. White fur is caused by the KIT gene, but it is not all-or-nothing. As you can see, Excuse is more than 50% white, which indicates that he is probably homozygous for the dominant white gene (SS).
Lastly, although it’s not clearly visible in this picture, Excuse’s dark patches have a tabby pattern to them. This is caused by the agouti and tabby genes, where the agouti gene determines whether a cat has a solid color coat, and the tabby gene determines the exact pattern.
Emily goes into much more detail in her Twitter thread, explaining colorpoint coat patterns, cats with diluted colors and more. If you are wondering why your cat looks the way it does, this thread will tell you all you need to know.
But in the meantime, what colour is your cat's coat — and do you know why?
Rapid analysis shows that the 2019-nCoV coronavirus resembles viruses from bats
Another publication suggests that this coronavirus enters cells in a similar manner to SARS
The 2019 novel coronavirus (2019-nCoV) outbreak has sparked a speedy response, with scientists, physicians, and front-line healthcare professionals analyzing data in real-time in order to share findings and call out misinformation. Today, The Lancet published two new peer-reviewed studies: one which found that the new coronavirus is genetically distinct from human SARS and MERS, related viruses which caused their own outbreaks, and a second which reports clinical observations of 99 individuals with 2019-nCoV.
The first cases of the coronavirus outbreak were reported in late December 2019. In this new study, Nanshan Chen and colleagues analyzed available clinical, demographic, and laboratory data for 99 confirmed coronavirus cases at the Wuhan Jinyintan Hospital between Jan 1 to Jan 20, 2020, with clinical outcomes followed until 25th January.
Chen and colleagues reported that the average age of the 99 individuals with 2019-nCoV is around 55.5 years, where 51 have additional chronic conditions, including cardiovascular and cerebrovascular (blood flow to the brain) diseases. Clinical features of the 2019-nCoV include a fever, cough, shortness of breath, headaches, and a sore throat. 17 individuals went on to develop acute respiratory distress syndrome, resulting in death by multiple organ failure in 11 individuals. However, it is important to note here that most of the 2019-nCoV cases were treated with antivirals (75 individuals), antibiotics (70) and oxygen therapy (75), with promising prognoses, where 31 individuals were discharged as of 25th January.
Based on this sample, the study suggests that the 2019 coronavirus is more likely to affect older men already living with chronic conditions – but as this study only includes 99 individuals with confirmed cases, it may not present a complete picture of the outbreak. As of right now, there are over 6,000 confirmed coronavirus cases reported, where a total of 126 individuals have recovered, and 133 have died.
In a second Lancet study, Roujian Lu and their fellow colleagues carried out DNA sequencing on samples, obtained from either a throat swab or bronchoalveolar lavage fluids, from eight individuals who had visited the Huanan seafood market in Wuhan, China, and one individual who stayed in a hotel near the market. Upon sequencing the coronavirus’s genome, the researchers carried out phylogenetic analysis to narrow down the virus’s likely evolutionary origin, and homology modelling to explore the virus’ receptor-binding properties.
Lu and their fellow colleagues found that the 2019-nCoV genome sequences obtained from the nine patients were very similar (>99.98% similarity). Upon comparing the genome to other coronaviruses (like SARS), the researchers found that the 2019-nCoV is more closely related (~87% similarity) to two bat-derived SARS-like coronaviruses, but does not have as high genetic similarity to known human-infecting coronaviruses, including the SARS-CoV (~79%) or Middle Eastern Respiratory Syndrome (MERS) CoV (~50%).
The study also found that the 2019-nCoV has a similar receptor-binding structure like that of SARS-CoV, though there are small differences in certain areas. This suggests that like the SARS-CoV, the 2019-nCoV may use the same receptor (called ACE2) to enter cells, though confirmation is still needed.
Finally, phylogenetic analysis found that the 2019-nCoV belongs to the Betacoronavirus family – the same category that bat-derived coronaviruses fall into – suggesting that bats may indeed be the 2019-nCoV reservoir. However, the researchers note that most bat species are hibernating in late December, and that no bats were being sold at the Huanan seafood market, suggesting that while bats may be the initial host, there may have been a secondary animal species which transmitted the 2019-nCoV between bats and humans.
It’s clear that we can expect new findings from the research community in the coming days as scientists attempt to narrow down the source of the 2019-nCoV.
Mini-brains are stressed out and not as reliable for disease modelling as scientists thought previously
Scientists compare cortical organoids to a developing human brain and spot key differences
You may have heard of organoids (probably here on Massive Science) and how they are the next big thing in science, especially when it comes to studying the brain. Sure, cerebral organoids are pretty cool. They're formed when stem cells self-assemble into 3D structures that recapitulate brain development, making organoids a good model to study development as well as disease. But now, on top of the ethical debates brought on by these organoids, new research out today suggests that they may not be as reliable as the field thinks.
A new study published in Nature thoroughly characterized lab-grown organoids (developed from human cortical cells) by looking at which genes they expressed and compared them with a developing human brain. They found that, compared to the creation of very specific cell types seen in normal developing human brain, organoids had broader cell types. What seems to be missing is the cell sub-type specification. Imagine a tree that grew a generic "fruit" rather than a specific thing like an “orange” or an "apple."
The organoids also seemed to be more "stressed out" than their normal counterparts. Cellular stress markers that had previously been associated with blocking development of those specific cell types were over-activated in the organoids, but not in the human developing brain. Interestingly, transplanting the organoids into mouse brains blocked the increased cellular stress, and also increased cell type specification in the organoids.
Although this may seem as an unfortunate turn of events, it does not mean that all organoids should be disregarded, but rather that scientists should be more wary when drawing conclusions. As statistician George Box famously said, “All models are wrong, but some are useful.” Knowing these limitations, the field must think carefully about the questions they want to answer using the organoid as a model.
Scientists built a 4D map of cell division
Cell division involves around 600 different proteins — it could take five or so years to study a single protein
Umberto Salvagnin via Flickr
Studying biology at a molecular level is often really hard.
Biological processes require the coordinated efforts of hundreds of proteins — the molecular machines encoded by our genetic material. Each protein needs to engage in a particular interaction or reaction at a very specific place and time. It’s complicated.
For example, mitosis, the process by which a cell divides into two, involves around 600 different proteins! Trying to sort out where each needs to be at a given time, in what quantity, and why, is an arduous task.
Thanks to a new technique developed by scientists at the European Molecular Biology Laboratory (EMBL), which combines a quantitative microscopy method with various computational analyses, our job just got a little easier.
This method can produce a 4D (that’s 3D plus time!) map of every single protein involved in a given process, and is first being applied to mitosis. Here's an example:
You can check out the resulting computer model, called the Mitotic Cell Atlas, now — simply choose any combination of proteins from those available (so far) in a drop-down menu to see how they interact at every stage of mitosis.
It’s still going to take several years to gather data for all of the roughly 600 proteins involved in mitosis, but it’s a huge step forward for efficiently analyzing protein dynamics.
Even better, this technique can be applied to many fundamental biological processes, which will certainly aid our understanding of how these processes go wrong in human disease and in other organisms. For example, scientists are already considering building a Plant Cell Atlas. Goodbye snapshots, hello movies!
You can add almost anything to improve graphene's function, even bird poop
For several years, researchers have been excitedly looking for the best dopant — before realizing that anything can improve graphene's electrocatalytic rate
By CORE-Materials on Flickr
Electrochemistry is the study of how chemical reactions generate electricity and conversely, how zapping solutions can trigger chemical reactions. Graphene, a mere sheet of carbon atoms, is known to be an excellent catalyst of electrochemical reactions as it can conduct electricity and has a large surface area to host reacting molecules. Furthermore, introducing dopants — impurities that can alter a material's physical properties — into graphene can boost its electrocatalytic activity. For several years, the research community has been excitedly looking for the best dopant that could lead to the greatest electrocatalytic improvement in graphene.
It was one of those promising, success story-filled research areas, where newer and more complex dopants would give rise to ever-improving electrocatalytic rates — until scientists realized that almost anything will improve the electrocatalytic rate of graphene.
These researchers laced graphene with guano, a low-tech fertilizer made of bird poop with usage dating back to the Incan empire. Compared to clean graphene, bird-poop laden graphene facilitated a higher rate of electrochemical reactions, typified by the oxygen reduction reaction (the reaction of dissolved oxygen) and the hydrogen evolution reaction (the breakdown of water).

The nest of the Peruvian booby is made of almost pure guano. This was found at La Vieja Island in Peru.
The reason for this enhanced electrocatalytic ability is that bird poop contains a hodgepodge of elements, including metals such as cobalt, manganese, and nickel. Basically, the more impurities graphene contains, the better its electrocatalytic reactivity overall. Call it quantity over quality.
In the paper, the authors lament the pursuit for the ideal graphene dopant, admitting such endeavors are “meaningless” and “never-ending”, and at one point, say:
"One may exaggerate only a little by saying that if we spit on graphene it becomes a better electrocatalyst."
According to the researchers, at least in the context of electrocatalysis, we don’t have to worry about “[making graphene] great again.”
On Holocaust Remembrance Day, the stories of two Jewish scientists
There were over six million Jews, allies and collaborators who were persecuted and murdered by the Nazi regime during the Holocaust
Today is the International Day of Commemoration, in memory of the victims of the Holocaust, where over six million Jews, allies and collaborators were persecuted and murdered by the Nazi regime. Six million is a difficult number to even begin to comprehend, so picture it this way: about two out of every three Jews in Europe were killed in this horrific genocide.
For many Jewish scientists, to practice science publicly in a region ruled by Nazi ideology was a death sentence. No one was exempt from this persecution. These are just two stories of those lost.
Leonore Rachelle Brecher — a skilled botanist
Born in Romania, Leonore Rachelle Brecher lived in a vibrant Jewish communities. At age twelve, she went to live with relatives after the death of both of her parents. Brecher went on to study zoology at a university in Chernivtsi, in what's now Ukraine. As her career started gaining traction, she moved to study at the University of Vienna where she defended her doctorate on color adaptation in insect chrysalides.
Applying to her alma matter, Brecher was denied a professorship. She later found employment in Berlin, and then moved through many universities and cities, before being forced back to Vienna in 1930. After the German occupation of Austria, she lost her job at the Vienna Institute for Experimental Biology because of her Jewish faith and began to teach in a Jewish gymnasium.
Like many Jewish European scholars, Brecher applied for refugee scholarship but was denied financial assistance by a British agency as her specialty was deemed too narrow, and rejected by the US because of the limited quota for Romanian immigrants. On 14 September 1942, Brecher was arrested and deported to the Maly Trostinets extermination camp, where she was killed immediately upon arrival.
Ernst Julius Cohen - a decorated Dutch chemist
Ernst Julius Cohen was born in March 1869 in Amsterdam, Netherlands. He studied Latin and Greek, before going to pursue a PhD in chemistry, under the supervision of J.H. Van't Hoff, at the University of Amsterdam, where his doctorate was titled "Het bepalen van overgangspunten langs electrischen weg en de electromotorische kracht bij scheikundige omzetting" (Determining transition points by electric way and the electromotor force in chemical conversion). After his defense in 1893, Cohen continued as a researcher, where subsequent positions included serving as a professor of physical chemistry in Amsterdam, and later, the director of a chemical laboratory at the University of Utrecht (until his retirement in 1939).

Ernst Julius Cohen (1869-1944)
After contributing to the discovery of two tin allotropes (white and gray tin), Cohen was propelled into the spotlight and began lecturing across the US and Europe. Cohen's studies spanned various fields, including electro- and thermochemistry, and was considered as the "greatest of the disciples of Van't Hoff."
Just like Brecher however, Cohen was caught in the Nazi's sweep across Europe. In 1941, Cohen's property was seized. In May 1942, Cohen was obligated to wear the "yellow star" and faced multiple restrictions because of his Jewish faith. In 1943, Cohen was arrested in his own laboratory on the charge of entering a "public" building. His friends' efforts were insufficient to secure his release, and he was deported to a concentration camp at Vught in Holland, but was fortunately released thanks to a plea made by the Council of the Dutch Chemical Society to the S.S. authorities at the Hague. However, troubles slowly continued, and on March 3rd 1944, Cohen was transported to the Auschwitz concentration camp, where he was murdered in the gas chambers.
Swamp sparrows can guess each other's ages from the sounds of their song
Researchers hand raised a flock of swamp sparrows, and used music playback experiments to see how wild males responded to different songs
By ethan.gosnell2 on Flickr
You pick up the phone and immediately categorize the voice on the other side — whether it's high pitched or deep, smooth or irritating, or if there's perhaps an accent or a quirky pronunciation. Magically enough, you can also estimate quite accurately if it’s the voice of a young, middle-aged, or older person. It turns out this phenomenon of guessing someone’s age by their voice is not exclusive to humans. In fact, swamp sparrows also do it.
Matthew Zipple, a biology graduate student at Duke University, set out to test a couple of hypotheses: whether older birds sing differently than younger birds, and if wild birds can spot these differences.
Firstly, it’s extremely difficult to determine a bird’s age in the field. Secondly, comparing young and old birds meant comparing completely different voices, not just different ages. How can you isolate the effect of age alone?
Enter a special flock of swamp sparrows (Melospiza georgiana), hand raised and followed by Duke University biologists Stephen Nowicki and Susan Peters.

A swamp sparrow sitting on a branch
By Andrew Weitzel on Flickr
Using songs recorded throughout these bird’s lives, Zipple and his collaborators showed that male songs peak in quality when birds are around two years old, then deteriorate over time, acquiring something of an old age signature. Older birds have less consistent songs and sing at lower rates than their younger counterparts.
Zipple and his team then presented these songs, young and old, to wild male birds. To do that, they placed speakers inside male birds’ territories and waited to see if the resident male would get upset over this intrusion.
These results, published in Behavioral Ecology in September 2019 and January 2020, demonstrate that humans are not the only ones who can tell age just by listening to someone’s voice. They also show that older birds seem to be less of a threat to other males, which is likely to mean that they are not as attractive to females as their young selves.
Fewer mates, but less aggression? Sounds like an okay way to grow old.
Be cautious about misleading coronavirus news. Here's how to spot what's accurate and what's dodgy
Incorrect reports spread like wildfire during events like these, so be prepared
muffinn via Flickr
In light of the recent coronavirus outbreak, new developments have been popping up every few minutes, especially on social media. However, everyone should be aware of misleading news and false reporting. While some claims are comically outlandish, such as the one about Bill Gates supposedly creating the coronavirus, other claims that misreport the coronavirus death toll or official WHO statements are more dangerous.
Here are a few things to keep in mind during this outbreak:
- Get your news straight from the source, such as your local and federal health organizations. This includes the WHO or the CDC.
- If a website is making a claim that you can't quite wrap your head around — check their sources! If they are citing an older version of their own article, that's a pretty big red flag.
- Be critical of the information you see, and always cross-check the consistency of facts by looking for the same information on different news platforms.
- The good old "correlation vs causation" rule. Yes, there is a video out there that shows someone eating a cooked bat. And yes, coronaviruses are known to infect bats. Does that mean that this video shows an example of an incident that caused a coronavirus infection? Not necessarily.
For more ways to dodge fake news, here is another great article by from Harvard University.
Also, remember, if you're feeling panicky about coronavirus, it's not nearly as worrisome as regular, old-fashioned influenza*, which we live with every day.
(Ed: We wrote this note in late January, and now in March it is obvious that we were wrong. to say this. Coronavirus is obviously in no comparable in severity to the seasonal flu. As editor I take full responsibility for this lapse in judgement. Aside from this note, I feel it would be unethical to change this piece to save face, so I am leaving it unedited for posterity's sake - Dan Samorodnitsky.)
Whales molt, and scientists are trying to work out how
Researchers examined tissues from bowhead whales
By Olga Shpak at Wikimedia Commons
Did you know that whales molt? They exfoliate by rubbing against rocks every spring. New research shows that whale molting has an underlying molecular process which may involve structures called desmosomes.
Desmosomes form between cells in the outermost layer of skin and are one of several factors that allow skin to be a flexible fortress, capable of defending the body and repairing wounds.
But these spots of glue do more than hold our skin together. They also hold together pumping heart tissue, and they keep the lining of our guts, kidneys, and various other internal organs intact, too.
Unlike glue which hardens, the desmosome is dynamic. It assembles, changes shape, or disassembles in response to various cues, including during cell differentiation, injury and wound healing. The desmosome is actually a complex formed by thousands of copies of five different proteins, which are intricately organized into discrete spots lining adjacent cells.
Most desmosome studies to date have focused on human and mice, but these structures are necessary for just about any organism that has skin. Without desmosomes, skin would break and blister very easily, giving foreign bodies easier access, as is the case in people with genetic diseases caused by mutations in the genes that encode the desmosome proteins.

A bowhead whale spyhops off the coast of western Sea of Okhotsk.
By Olga Shpak at Wikimedia Commons
This new research on whale molting mechanisms suggests that desmosome loss can also be functional. Researchers took tissue samples from bowhead whales (Balaena mysticetus) across three different sub-groups: molting adults (spring), non-molting adults (fall), and molting juveniles (spring). Next, they used a technique called immunohistochemistry to test for the presence of two desmosome proteins: desmoglein and desmocollin. The levels of these proteins have been previously shown to regulate desmosome assembly and adhesion.
The researchers found that there were season-dependent decreases in the levels of desmocollin, consistent with skin shedding. For example, in all spring-caught molting adults, there were reduced desmosome proteins in their oral cavity.
This study suggests that skin shedding in mammals, whether it's a result of normal molting or is the result of injury or disease, involves desmosome function. As for bowhead whales, more work is needed to understand the mechanism, and to determine if these changes in desmosome protein expression occur in other organisms with annual molts.
Researchers develop an app for tracking trade of endangered fish
Say "cheese!" for Napoleon wrasse conservation
Larry Koester on Flickr
The Napoleon, or humphead, wrasse (Cheilinus undulatus), is a giant and ornately decorated reef fish which can grow over 6 feet long and live for over 30 years. Unfortunately, these rare fish are a coveted delicacy in live fish markets throughout Asia, and the high volume at which they are traded has landed them a spot on the CITES endangered species list.
To keep them from going extinct, stricter regulations must be implemented and enforced surrounding the collection and trade of Napoleon wrasses. Unfortunately, fish trading networks are complicated, and it can be difficult to track where fish come from and if they are being sold legally.
While all fish imported to major markets may be inspected, those within the live fish trade cannot easily be tagged or marked, making it difficult to track these fish from the point of capture through to their final sale point. But a pair of researchers from the University of Hong Kong decided to try a new technique in lieu of a physical fish barcode: facial recognition software. Napoleon wrasses have complex facial patterns that the researchers hoped could be used as a unique identifier for individuals.
First, they had to test if the facial markings were unique and variable enough between fish to reliably identify individuals, and that they did not change over time. Then to ensure that the identification system was feasible, researchers developed and tested an app used to compare pictures of Napoleon wrasses at fish markets to those in a database of previously identified fish. And it worked – this study served as a proof-of-concept that facial recognition software could reliably identify endangered Napoleon wrasses sold in live fish markets.
Although not currently used on a large scale, there is real potential that the software and app developed could improve the monitoring and enforcement of legal endangered species trading. This approach could also apply to other animals that have distinct, readily identifiable markings.
Puffins captured on film using sticks as a tool to scratch themselves
It's the first observation of seabirds using tools
Photo by Guilherme Romano on Unsplash
Zoologists have captured footage of puffins performing an unexpected behavior: using a wooden stick to scratch a difficult-to-reach itch. Analysis of animal behavior is important for understanding both evolution in the broad sense but also the evolutionary history of humans. One behavior humans excel at is tool use and understanding the origins of this behavior also gives us insight into what it means to be "us." Although tool use has been documented in some other species — like for food extraction in apes and crows — this is the first time body-care tool use has been recorded in seabirds. The researchers found two independent instances of puffins using a stick for body care in locations more than 1,700 km apart.
This indicates a higher cognitive function in the seabirds than previously thought, and begs the question whether all puffins have this ability or if it arose in certain puffins only as an adaptation to the environment. Above all else, this finding emphasizes that tool-use may be more widespread than formerly believed and a better understanding its origins may require the inclusion of additional, understudied taxa.
My brother has 4% Neanderthal DNA. What does that actually mean?
Neanderthals are an extinct relative of Homo Sapiens, having died out around 40,000 years ago
Photo by mostafa meraji on Unsplash
Like a lot of Americans, my brother decided to fork over some cash and have his DNA analyzed to see his ancestry via 23andMe. The usual information popped up — some German ancestry, some Scandinvian, but then a strange name and number...4% Neanderthal.
Neanderthals are an extinct relative of Homo Sapiens, having died out around 40,000 years ago. They’re not exactly roaming the streets of Stuttgart like some of the other Vogts I’m related to. But, surely there are some applicable traits that we share, right? After all, my brothers and I often joked about our physical anomalies while growing up — we have large rib cages and shoulders, wide nostrils, and can handle brutal cold temperatures, which are all traits of what scientists such as Joshua Akey have attributed to Neanderthals.
What this DNA does define, according to The Scientist, is that my modern human ancestors bred with Neanderthals roughly 50,000 to 55,000 years ago. A variety of scientists working on genome sequences have come up with different concepts as to why humans mated with their closely related cousins, and the effects it has on their far-flung descendants, such as my family. One of them is the effect on skin, as well as fatty tissue beneath, as Neanderthals survived in brutally cold environments thousands of years ago. Finally, a study mentions that some of these genomes may relate to psychiatric disorders that are prevalent in some human populations, which I myself happen to have.
Nothing much outside of interesting facts as they come, but I do find it funny when certain individuals glorify blond, blue-eyed, fair-skinned individuals like myself and my brothers. They have no idea that we’re not fully human. Maybe we should take them back to our caves and teach them how to get along, Neanderthal and human alike.
Solitary confinement is bad for the heart too
A new study shows that a third of individuals in solitary confinement are more likely to have heart attacks and strokes
By Mitch Lensink on Unsplash
Solitary confinement does little to rehabilitate inmates, is extremely expensive (where the average per-cell cost is $75,000), and exacerbates health problems — yet the American prison system is over-reliant on solitary confinement. In fact, a 2018 report found that, 61,000 individuals were being held in solitary confinement across the US. While the living conditions in such units are known to be associated with adverse health outcomes, we still don't understand the lifetime cardiovascular burden (i.e. the direct and indirect costs) associated with solitary confinement.
To explore the health consequences of solitary confinement, Brie Willams and her colleagues compared the lifetime cardiovascular health burden from solitary confinement, relative to those of inmates who were not held in isolation.
Hypertension refers to abnormally high blood pressure.
By jesse orrico on Unsplash
The researchers first looked into public data from a 2015 lawsuit. This data contained descriptions of hypertension diagnoses among two groups of men: individuals in the first group who were held in a prison’s solitary confinement “supermax” units (the “Supermax Unit Group”), while individuals in the second group were housed in regular, less-isolating maximum security units (the “Less Isolated Group”). Both groups’ levels of loneliness were measured with the UCLA Loneliness Scale.
The researchers then used the Cardiovascular Disease Policy Model to estimate and compare the lifetime burden of cardiovascular problems for both groups, while accounting for potential factors, such as socioeconomic factors, which may affect comparisons. Medical costs associated with the cardiovascular disease burden were also estimated using California’s health costs, and these estimates were then deflated to the U.S. national average costs.
The study found that the hypertension rates were 31% higher in the SuperMax Unit Group than those in less-isolated units. In addition, about a third of the SuperMax Unit Group were more likely to experience heart attacks, strokes, and higher scores of loneliness.
In fact, the authors point out that if their findings were applied to simply 25,000 individuals held in “supermax” solitary confinement units, this alone would result in $155 million in additional future healthcare costs — and would likely still be an underestimate as there is a widespread use of solitary confinement beyond “supermax” units.
These findings call for a reform in American prisons. Evidence already shows that solitary confinement is counter-productive — this is one more study which points to the health consequences of solitary confinement.
Specialized nerves let squid tentacles strike with lightning speed
Squid have different types of nerves in appendages with different functions
NOAA Ocean Exploration & Research
Contrary to octopuses, which only have arms, squid have both arms and tentacles. You can distinguish between the two by looking at the tips: tentacles have a bulb at the end, called a club, but arms do not. Additionally, the tentacles are extendable while arms aren’t, reflecting the difference in function between the two. Squid use the tentacles to strike at their prey and reel them in, at which point the arms take over and subdue the prey.

Squid have arms AND tentacles. Here, the tentacles are the whiter appendages with the clubs at the end.
prilfish via Flickr
Squid can extend their tentacles in 20-40ms, which is about 10 times faster than a blink. This is especially impressive, since they don’t use any spring-like mechanisms. They only use their powerful muscles for this, which are arranged in a special criss-cross pattern. However, their muscles aren't the only things with special adaptations.
On 3 January, William Gilly from the Hopkins Marine Station and his colleagues published a study where they studied the nerves that drive these muscles. The most striking difference they found between the nerves that innervate the arms and those that innervate the tentacles, is that only the latter could produce action potentials. Action potentials are extremely fast electrical events where the inside of a nerve cell temporarily becomes more positive than outside the cell. This may cause the release of neurotransmitters, which in turn causes the muscle to move.
Action potentials are widely used in vertebrates, but invertebrates, including the squid, often use graded potentials. This means that rather than a fast electrical event, there are gradual changes of the electrical charge. And this is the type of communication that the nerves in the arms of the squid use.
By using the quick action potentials for the tentacles, the squid can perform rapid strikes to catch their prey. The arms don’t need to move as quickly, so no action potentials are needed to move those. This is how squid not only evolved the muscles in their limbs, but also the nerves, to optimize each type of limb for its own specific job.
How many things can scientists name -Seq? Let us count the ways
From lettuce to cat poop to human cells, we're ready to sequence it all
In the past decade, scientists have been slowly sequencing everything — and yes, I mean everything. Scientists have sequenced romaine lettuce to detect food-borne pathogen outbreaks, increasing numbers of people across the world to address under-representation in genomics, and even kitty poop to better understand the cat microbiome.
But in the midst of all these sequencing studies, what sticks out to me the most is how often scientists find a way to use the word -Seq when naming new methods. Here, Seq is short for sequencing, and is often used to name experiments which involve high-throughput sequencing.
So here's a list of the many ways scientists have managed to incorporate the word -Seq when naming new methods. It certainly isn't comprehensive, and doesn't include every -Seq method out there, but here it is anyway.
Here are (some of) the ways you can sequence DNA

The Oxford Nanopore MinION is a pocket-sized DNA sequencer — just in case you want to sequence on the go.
There are so many different aspects of biology to consider today when it comes to sequencing DNA. Are you interested in learning what DNA interacts with? Try using ChIP-Seq to explore protein-DNA interactions, or if you want to get more specific, use MAINE-Seq to find DNA bound by histone proteins.
Interested in the regulatory regions of the genome? There's also FAIRE-Seq to identify regulatory regions in the human genome, and 4C-Seq to characterize how DNA is physically organized around regulatory elements. Or perhaps you're interested in whether DNA is methylated or not? If so, you can take your pick between BS-Seq, BisChIP-Seq, Methyl-Seq and so many more.
Want to look at RNA? There's a -Seq for that, too
The first step to building a protein in your cells is to transcribe DNA into RNA (ribonucleic acid). Scientists can map RNA using RNA-Seq, but that's not all. Check out RIP-Seq to find out where RNA-protein complexes are, dsRNA-seq to investigate double-stranded RNA molecules, and structure-Seq to find secondary RNA structures across the genome. There's even single-cell RNA sequencing (scRNA-seq) if you want to look at individual cells, which was hailed as Science's Breakthrough Of The Year in 2018. Fun fact: scRNA-seq was originally called mRNA-Seq, but perhaps that name wasn't catchy enough.
Don't forget about CRISPR!
Scientists can use the CRISPR/Cas system to edit genes, but it doesn't always work out effectively. So why not use GUIDE-Seq to identify all the double-stranded breaks introduced by CRISPR/Cas enzymes? Or Digenome-Seq to capture off-target CRISPR effects in human cells? Or perhaps CIRCLE-Seq to capture off-target effects in vitro? Or...okay, I'll stop now. But there are so many more -Seq methods out there as the scope of sequencing continues to expand.
But let's face it: I also carry out DNA sequencing to better understand complex neurological disorders. If I ever developed a method, I'd try to find a way to fit -Seq in too when naming it. How else would you know that sequencing was involved?
Why did blue whales get so big when their prey is teeny tiny?
When the food source is abundant, animals with bigger mouths can consume more calories.
NOAA Photo Library via Flickr
When we think of large animals, whales immediately come to mind. Whales come in all sizes, though. Why aren’t dolphins and orcas (which are technically toothed whales) as large as blue whales? And how come blue whales, the largest of all animals, survive on the tiniest of prey items, such as krill? A new study published in Science explores these questions.

Amanda Slater via Flickr
Being so large could help whales dive deeper and for longer, which could be good for finding large prey. Yet, for toothed whales, such as sperm whales and porpoises, such large prey is rare and hard to find. This means that, although larger toothed whales can dive deeper, their prey/dive ratio is smaller than that of smaller whales. For baleen, filtering whales, however, the story is different. There is plenty of krill in the ocean. One of the main constraints to eating, for baleen whales, is to open their mouths wide enough to swallow massive amounts of krill-containing water. And guess who has bigger mouths? Bigger baleen whales.
For baleen whales, feeding efficiency increases with mouth size, which in turn increases with large body sizes. Huge baleen whales can swallow more calories per dive than their smaller cousins. Why aren’t blue whales even bigger, then? Scientists think that it is probably due to seasonal shortages in krill density. During dry spells, fat energy storages would probably not be enough to sustain an even bigger whale.
For toothed whales, on the other hand, being very large is not really an advantage, since there are very few large prey to be found in the ocean. It’s more energetically advantageous for these toothed predators to be medium size and eat medium size prey, than to dive long and deep searching for an elusive large meal.
Go to bed: sleep deprivation changes how you experience pain
New research highlights the role of the nucleus accumbens in how sleep-deprived individuals respond to pain
We have all felt the sensation of pain. And we are also all familiar with sleep, a necessary bodily process, has cognitive and physiological functions. While sleep is behaviorally regulated, research shows that poor sleep can actually tamper with our brain's processing of pain. Using fMRI imaging, scientists have now visualized the amplification of the pain signal in the somatosensory cortex of sleep deprived individuals.
The somatosensory cortex is the area of the brain that registers pain stimuli, meaning it tells us where the pain sensation we feel is coming from. Then, the pain signal moves into the insula, the part of the brain which integrates all incoming signals and creates our conscious perception of pain. Following that, the signal is sent along to the nucleus accumbens (NAcc) which plays a major role in decision-making, reward, and pain evaluation.
The study, published last year in the Journal of Neuroscience, found that the activity of the NAcc in sleep deprived individuals is altered: compared to the pain response in people who aren't sleep-deprived, the NAcc dulls incoming pain signals, increases pain-relief seeking behavior and affects decision-making.
A previous study highlighted the increase of opioid consumption in burn patients after just one night of interrupted sleep, which reinforces the idea that sleep and our sensations of pain are closely linked. While research strongly supports the idea that sleep deprivation does increase pain, but exactly how our brains process the interplay of sleep and pain is still being investigated. For instance, the NAcc has broad connectivity with many regions of the brain including the prefrontal cortex, anterior cingulate cortex, and the amygdala (just to name a few). Future research should tease apart the question of “What’s more important: the amount of sleep we get or the quality of sleep?”
But in the meantime, consider counting sheep an aid to resetting our bodies and our brains.
Scientists, beware of the consequences of routine practices in your lab!
The harmless practice of using parafilm to seal agar petri dishes containing the model organism, C. elegans, actually impacts larval development
By Michael Schiffer on Unsplash
What if some of your common practices in the lab were actually having a significant effect on your results...and you just didn’t know about it? Recently, a group of researchers from Utica College found that the practice of using parafilm to seal agar petri dishes containing the model organism, Caenorhabditis elegans, actually has a significant effect on its early development.
C. elegans is a nematode and a well-established model organism which is often used to model various aspects of development. This organism is often grown on Nematode Growth Media (NGM) agar in a petri dish.
Meet Caenorhabditis elegans — a common model organism used in biological sciences.
NGM can sometimes dry out or become contaminated by microbes, so to prevent this, researchers often wrap up the petri dish with parafilm, the way you wrap up food containers with plastic wrap in the kitchen. This practice has been used for a long time, however, it has never been fully explored to see if it actually affects nematode development. In this study, researchers tackled this very question and investigated whether wrapping NGM agar plates with parafilm had an effect on larval growth and development.
In their study, the researchers found that both the larval growth rate and length change after 48 hours were significantly increased when the NGM agar plates were wrapped with parafilm.
Researchers also found reduced variability in growth among parafilm-wrapped replicates in this study. This echoes findings of a previous study which looked at the effects of parafilm on the model plant, Arabidopsis thaliana. Overall, this suggests that parafilm helps to create a standardized condition for measuring responses in model organisms.
Any researcher that uses parafilm as common practice with their organism should be aware of these possible effects and take the time to identify if this practice of wrapping up has any influence on their results. And in addition, perhaps there are other types of common practices out there that can affect results...that we just don’t know about yet.
I chased lemurs around Madagascar to help stave off their extinction
Planting fruiting trees is a vital component of reforestation and conservation efforts
Brian Gratwicke on Wikimedia Commons
A lemur, a primate cousin that evolved separately on the island of Madagascar, leaps above me through the rainforest canopy. Ninety percent of the lemurs on this island off the east coast of Africa are in danger of extinction. I am studying the Milne Edward’s sifaka, an endangered species of lemur in Ranomafana National Park. I am following these lemurs around to better understand their diet and behavior, research which hasn't been updated in over a decade but is very important for this species' conservation.
I’m a senior student at Rice University collecting data for my honor’s thesis. I received a grant from my school to go to Madagascar to conduct this research.
I run through the forest after the lemurs, taking observations of their behavior every three minutes, following the protocol for past studies. During my time here I am following six different groups of lemurs, three in degraded forests and three in pristine forests. The group I'm following today, which occupies the disturbed forest, consists of two adults females and two juveniles. They run out of sight down a steep ravine. One false step, and I slide down the muddy slope on my butt. Desperate for every piece of data, I pick myself up and keep running to maintain pace with the lemur I’m focused on. She’s a mother, and a green-eyed baby pokes her head out below mom's belly. She nibbles on some young leaves, a common food item in this degraded forest.
As compared to the pristine forest, there are less vines and more tall trees with fruits and seeds. So far, my observations mirror the finding of previous research that lemurs eat more fruits and seeds in degraded forests because these foods are relatively more available. This research shows the importance of having these food items available for the sifaka lemurs to eat and reinforces the approach taken by reforestation efforts elsewhere on the island: tree species that yield abundant seeds and fruits should be prioritized.
I will return to the US after my time with the lemurs and publish these results. Hopefully, this will aid conservation managers in informing reforestation efforts to provide food for the endangered Milne Edwards’s sifaka lemurs.


















