
The soil microbiome could change agriculture—if we can understand it
Tinkering with the buzzing "social network" in the soil could be a new frontier for making crops grow faster and healthier.
If social media feels overwhelming these days, it’s nothing compared to the chatter going on beneath your feet. Pick up a handful of soil and you’ll be holding five times as many microbes—tiny, living organisms like bacteria—in the palm of your hand as there are people on Twitter. Like Twitter users, soil microbes have a lot to say about the world around them.
Microbes talk to each other by dropping small molecules like hormones into the soil around them. But they're not the only ones listening. Plants living in soil can interpret these messages as signals to grow out their root system, germinate seeds, or grow their stalks faster.
It turns out that the way soil microbes interact with each other has a huge impact on how plants grow. In fact, microbes can protect plants from disease and prevent plants from drying out during extended droughts. And yet research within the agricultural industry has largely focused on fertilizers, plant breeding, and genetically modifying plants. The billions of microbes that live alongside plants have received scant attention.
The hidden community in your backyard
As researchers have started to explore this world, they're realizing that we know far less about soil microbes than we thought. You might have heard of the human microbiome, the community of bacteria that lives inside every human. Soil microbe communities put our digestive systems to shame. Every spoonful of soil contains an estimated 100,000 unique microbial species, compared to less than 1,000 different species in the gut of any given human. In fact, over one-third of the microbial tree of life was discovered from soil only last year—before that, scientists didn’t even know that these microbes existed in soil.
Just knowing they exist doesn’t even scratch the surface of the all-important question: what are each of these microbes doing, and how do their activities affect plant growth?

Bacteria on the surface of the root of a fungus.
Image by BrothersOfMetal via Wikimedia Commons
That’s a huge unanswered question, with big consequences for food production. Global food demand is expected to increase by 35 percent over the next 15 years. In the same period, the amount of land available for agriculture is anticipated to decrease by 2 percent. While advances in plant breeding and genetically modified plants may help to close that gap, researchers are taking a new look at the potential of using soil microbes to crank up the output of existing farmland.
Building better soil
The idea of “engineering” microbial communities in soil has gained traction as a way to maximize the benefits of the microbial social network for crops. Microbial engineering—changing the composition of microbes that make up a community—has already shown promise for improving digestive health in the microbe-filled human digestive tract. There, microbial engineering relies on probiotics, which add desired microbes into the community, vitamin supplements, which help specific gut microbes grow, and even surgical fecal transplants, which transplant gut microbes directly from one person to another.
In soil, microbial engineering could be as simple as adding food sources or vitamins that beneficial microbes need, adding antibiotics that target detrimental microbes, or infusing fertilizers with probiotic cocktails of plant-friendly microbes.
There's a catch, however. Without knowing what each of the microbes in soil do it's impossible to decide which microbes to add or subtract to maximize crop yields. The microbial social network is not only beneficial to plants, but essential to the entire community of organisms living in soil. Microbes often cannot function without the support of their social network since they’re constantly interacting with their neighbors by trading food, removing toxic waste, and sending signals about changes in the soil around them.
Adding or removing the wrong types of microbes could upset the delicate balance of the community, with disastrous results for plant growth. As a result, scientists have to understand not just the microbes that directly affect plant growth and soil health, but all of the surrounding microbes that play an indirect role in supporting the entire microbial community.
Unfortunately for scientists, the interconnectedness of soil microbes also makes them nearly impossible to study in isolation in the laboratory, which is how we have typically figured out how different microbes work. Isolated from the other soil microbes they normally interact with, microbes brought into the lab simply die.
Hitting the side of the barn
Instead, soil researchers are turning straight to the source for answers: DNA. As the blueprint for life, the DNA in every microbe’s genome defines what it can do: what it eats, how it interacts with other microbes, and whether it provides any direct benefits for plants. If scientists can map the genomes of every microbe in the complex soil community, they can figure out which microbes are involved in plant growth and start to untangle the complex web of soil communication.
The key to mapping these genomes is a relatively new technique known as genome binning. Rather than attempting to untangle individual cells in the laboratory, scientists are mashing up all of the microbes in a spoonful of soil and sequencing all of the DNA that spills out at the same time. The process is known as “shotgun” sequencing for its molecular-scale resemblance to shooting birdshot at the broad side of a barn.
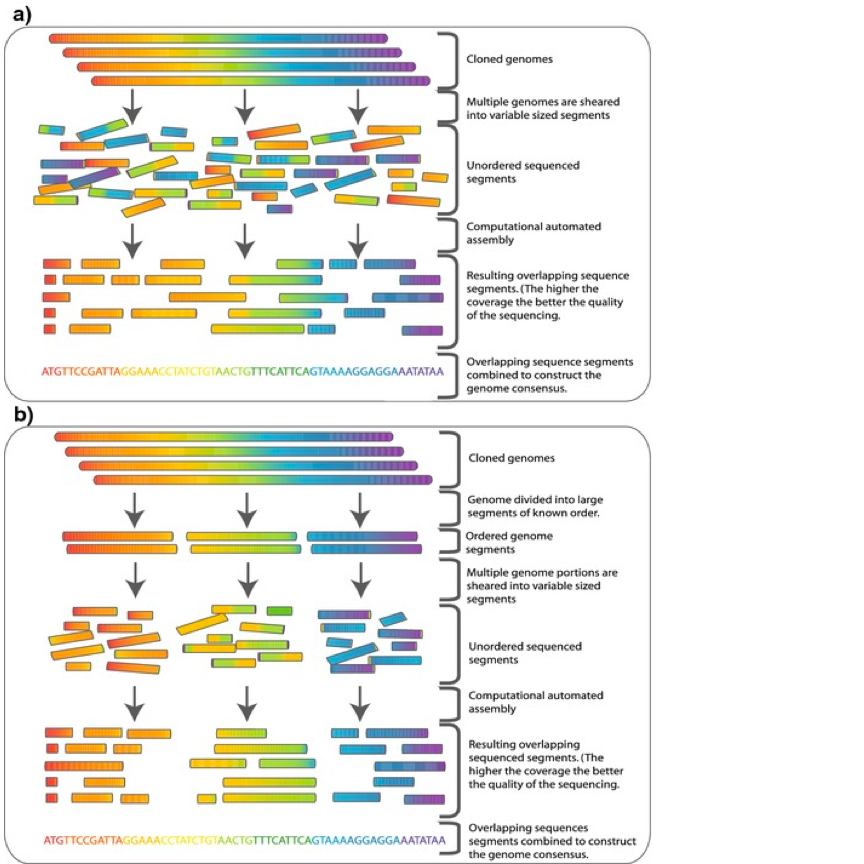
A diagram showing a shotgun sequencing process.
Illustration by Commins, J., Toft, C., Fares, M. A. via Wikimedia Commons
Shotgun sequencing results in millions of short, fragmented DNA sequences, each representing a small piece of the genome of one of the 100,000 different microbes in that soil. The problem is figuring out which of those fragments belongs to which microbe. Genome binning has been successful in simple microbial communities with only a few hundred different species. But until recently, success has eluded soil researchers because of the sheer number of microbes living in soil.
Rising to the ‘grand challenge’
At the heart of genome binning is a computer algorithm that has to decide which DNA fragments belong together and which don’t. To accomplish that, the algorithm compares every individual DNA fragment to find matching DNA sequences, which indicates that two samples generated by shotgun sequencing are actually consecutive sections of the same organism’s piecemeal genome. As the number of different organisms contributing to the pool of DNA fragments sequenced increases, however, so does the number of possible DNA sequences and misleading partially overlapping sequences. When there are too many different organisms present, the algorithm can’t confidently identify which overlapping fragments truly came from the same genome, so it fails to bin any genomes at all.
Because soil samples contain so many organisms, binning has become known as the “grand challenge” for soil scientists. And a group of researchers at Pacific Northwest National Laboratory led by Dr. Janet Jansson may be on the verge of tackling it.
To address the overlap problem, these researchers took a somewhat unconventional approach. Instead of trying to improve the computer algorithm that matches DNA sequences, they wondered what would happen if they could improve the DNA fragments themselves. Until recently, the best DNA sequencing technology available could only produce fragments of about 250 units long. These short sequences don't contain a lot of information, which means the chances of finding two sequences that seem to match, but don't actually belong together, is quite high.
But the latest DNA sequencing technology, released only three years ago, can sequence DNA fragments that are up to 10,000 units long, a 40-fold improvement over the next best technology. These long fragments make it much easier for the algorithm to find unique overlaps in DNA sequences from the same microbe, regardless of how many other microbes contributed DNA to the pool.
Dr. Jansson’s group put this new technology to the test by chopping up and shotgun sequencing the DNA in three spoonful-sized samples of prairie grass soil, which is the soil we rely on for the Midwestern US Corn Belt and the majority of the US’s agricultural production.
(NRCS_Photo_Gallery).jpg)
A scientist takes a soil sample in an Iowa corn field.
Photo by Tim McCabe, USDA Natural Resources Conservation Service, via Wikimedia Commons
The new sequencing technology returned more than 15,000 fragments, each at least 9,000 units long. When the team ran these long fragments through the binning algorithm, the computer returned 129 reconstructed genomes from microbes living in the prairie soil. That’s 129 microbes that scientists can now investigate to determine what roles they play in the microbial social network and how they may impact corn production.
Plowing through challenges
Despite the recent success, scientists still aren't done perfecting the shotgun sequencing and genome binning technique. For starters, the 129 genomes that Dr. Jansson’s team recovered are far from complete. In many cases, the genomes are estimated to be missing more than half of their DNA. That means that while researchers can gain some insight into what these microbes do based on their incomplete genomes, it will be impossible to gauge the full range of interactions the microbes participate in.
And even if scientists can iron out those issues, it may be a long time before it's economically feasible to study soil microbiomes on a large scale. Analyzing those three prairie soils ran up a bill of more than $6,000 thanks to the premium placed on the latest, proprietary sequencing technology. While the price of DNA sequencing continues to drop across the board, in the near-term that high cost may render this approach too pricey for small research groups or for important large-scale surveys that compare multiple types of soil.
Nevertheless, Dr. Jansson’s team demonstrated that untangling soil microbial DNA and understanding the soil microbial social network, the “grand challenge,” is achievable. While microbial engineering in soil to improve crop yields may not be around the corner just yet, this development elevates it from a farmer’s dream to something that can be actively pursued for globally important food crops. With a clear path forward for soil researchers, the day may be coming when we can decipher what soil microbes are saying to each other and harness that underground social network to re-shape the world's agriculture.
