
Photo by Fredrick Tendong on Unsplash
Gamers are one-upping nature and computers by designing whole new proteins
Building elaborate structures from scratch is difficult for computers, but in the hands of a gamer...
The idea of computer versus human has been intriguing. Humans have built computer algorithms that perform incredible feats – Google DeepMind’s AlphaGo and AlphaZero are mastering complex games of Go and chess, and defeating human champions faster than anyone thought possible.
But, there is a place where humans are teaching computers how to solve problems, not the other way around. One research group in Washington has asked the public to help them pursue one of the knottiest problems in biology: how do proteins fold?
In 2008, Professor David Baker’s lab at the University of Washington, with other collaborative institutions, developed and released FoldIt, a downloadable game accessible to anyone with a computer. The game would teach players the basics of protein structural biology, then challenge them to predict the structure a given simulated protein would fold into. Players can wiggle, shake, or swing the protein around to arrange it into its most stable configuration.
The players ended up excelling and going beyond just predicting structures. By 2011, they were helping stumped scientists solve the structures of real life proteins – for example a retrovirus protein (for a monkey virus similar to HIV). The following year, in 2012, they took an enzyme designed by scientists and modified it in FoldIt by rearranging part of the structure so it worked 18-times better.
The players then took it another step forward. Now, researchers have reported in a recent paper published in Nature Letters that citizen scientist gamers, by playing FoldIt, came up with their own design for stable protein structures from scratch.
The game is...weirdly addictive.
The authors of the paper, led by University of Washington research scientist Brian Koepnick, took 146 player-designed proteins from the simulated world of FoldIt and synthesized them in the lab. Out of these, 56 were stable in real life, with 20 different types of folds (like a helix or a bend), one of which has never before been observed in nature. When the researchers used biophysical techniques to obtain detailed 3D structures of four of the player-designed proteins, they saw that the real-life structures were pretty spot on with what the players designed on the computer.
Proteins are molecular machines responsible for running each and every cell that make up every single living organism on earth. Their functions come from their structures, in the same way that a hammer's ability to drive nails comes from the flat surface on its head. Proteins are made of amino acids, arranged with each one connected to the next like beads on a string. Amino acids, each with their own properties and preferences, can push and pull on each other to force the entire protein into a stable shape. But it’s hard to predict how amino acids connected in a 2D linear chain will arrange themselves into a 3D structure.
The human body alone is predicted to have at least 10,000 types of unique proteins. To experimentally determine even one protein’s structure in the lab is a long, tedious, and expensive process that isn’t even guaranteed to work. So we strive to be able to predict a protein’s structure with just its amino acid sequence without having to put in that time. Unfortunately, there’s no direct translation from amino acid to 3D structure. There are just too many possibilities for a chain of amino acids to arrange itself. Computers can simulate protein folding and run through the possibilities to find the most stable structure faster than we ever could.
If protein structures are hard to predict and harder to customize, they’re even harder to build from scratch. Consider predicting how a 2D linear chain of amino acids folds into a 3D structure versus having a 3D structure in mind and going backwards, predicting what amino acids you can use to design it. The possibilities are vast even for a computer to handle. Yet, everyday gamers learned to fold proteins in ways that set them apart from computer algorithms.
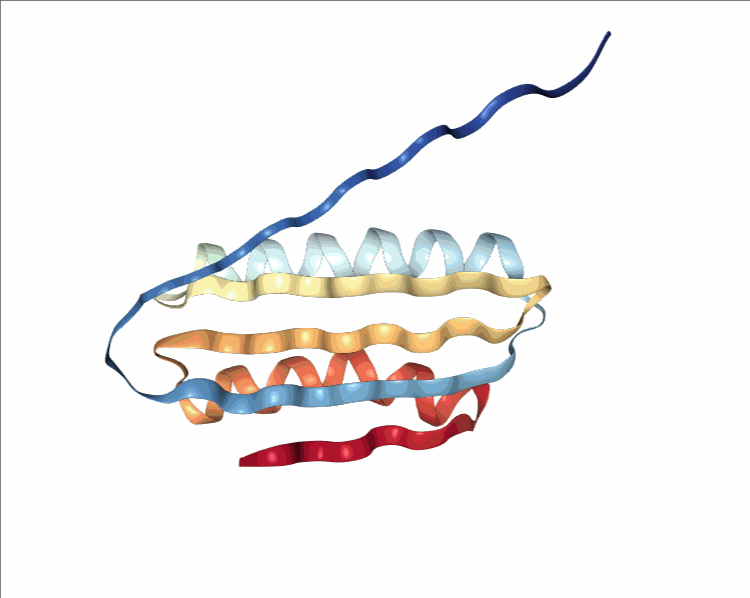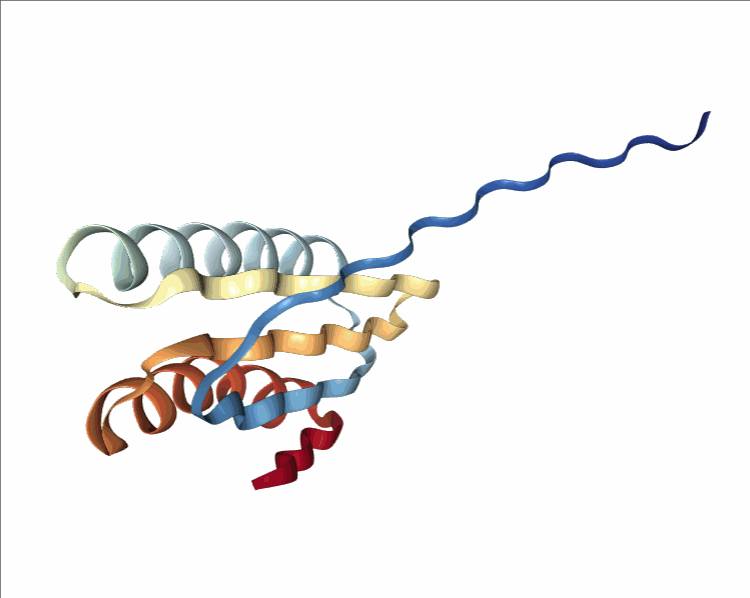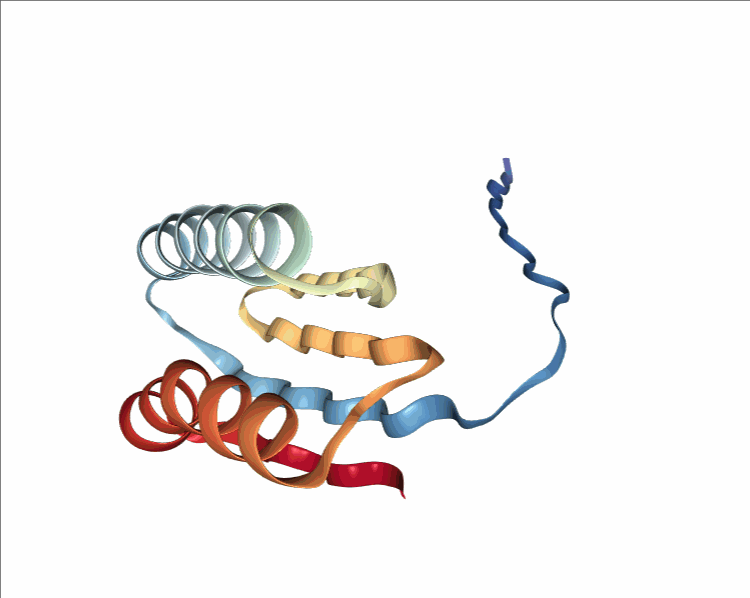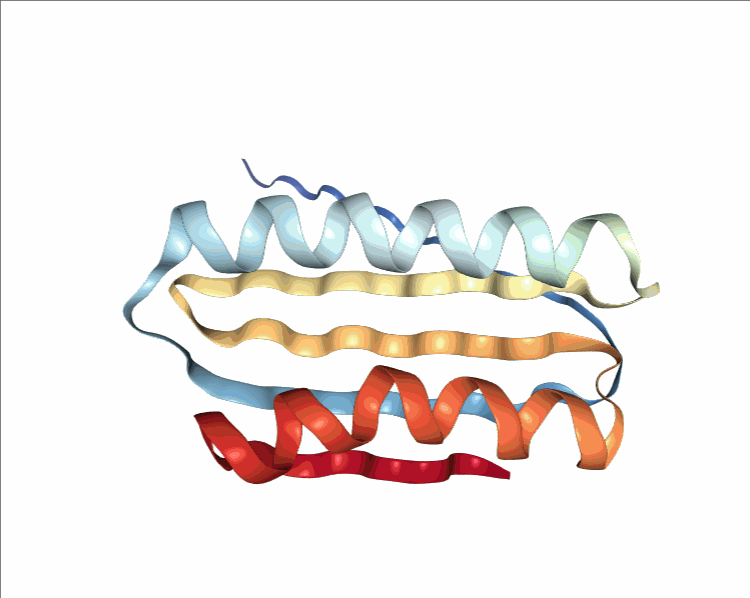
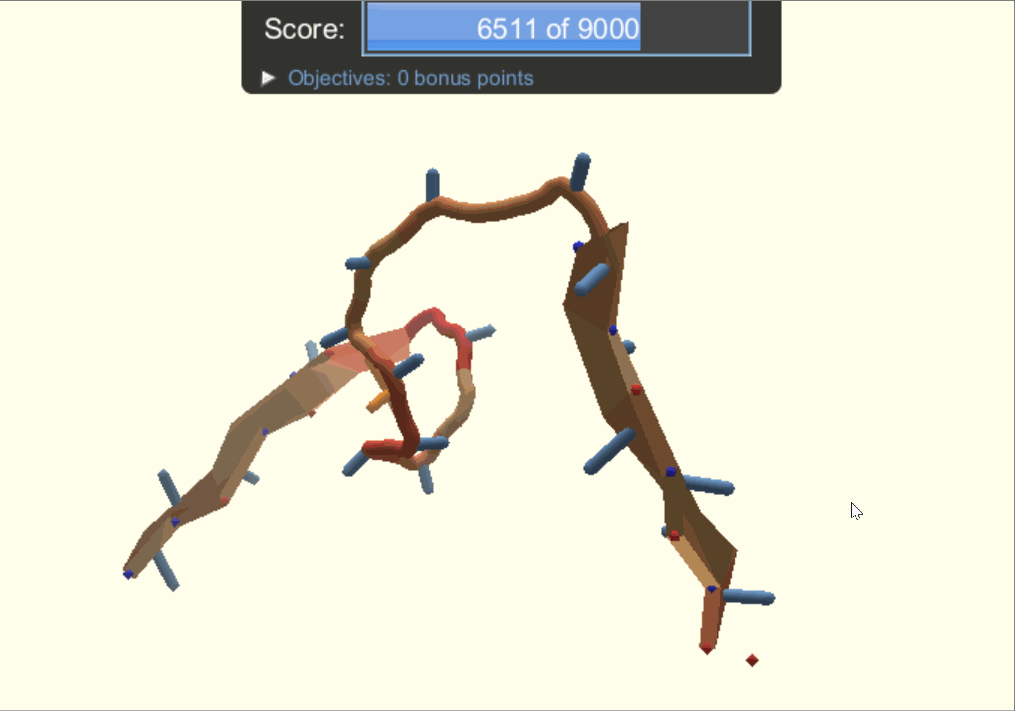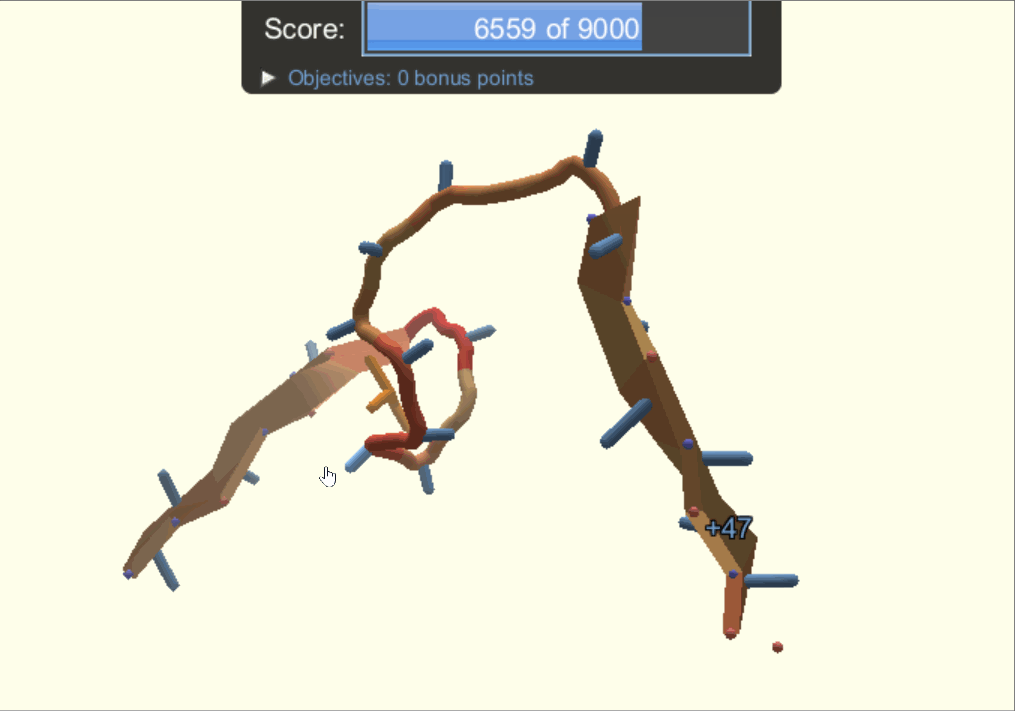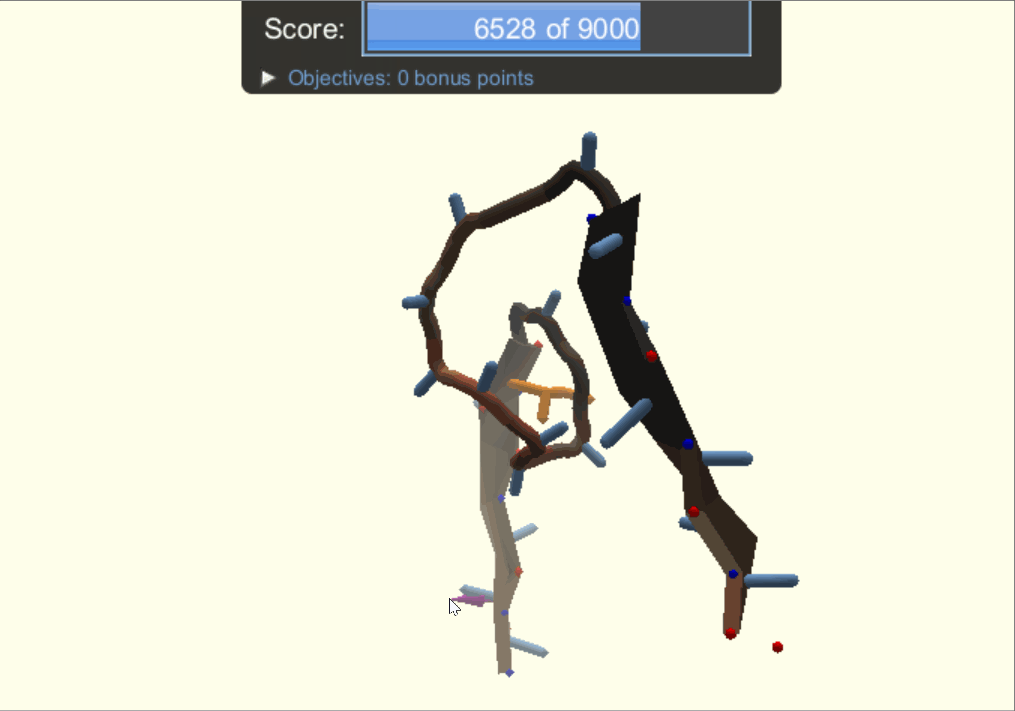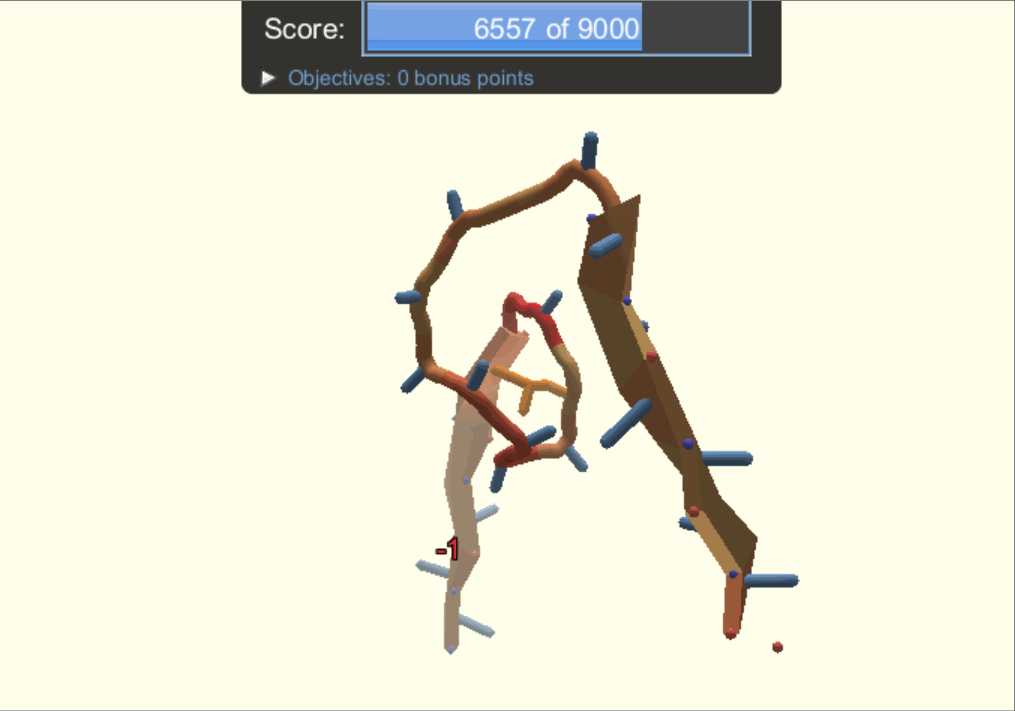
One of the player-designed proteins, called Ferredog-Diesel.
PDB: 6NUK
To understand the players' methods and strategies for designing proteins, the authors looked through snapshots of their structures throughout their design process. They noticed that players would frequently go back to earlier versions of their structure to try out other options of folding their protein. As a result, gamers end up exploring a complex variety of possibilities while most typical computer-automated programs would consider two options, make the decision, and move on.
This also meant that to arrive at a final stable structure, players tried out more unstable structures to try new possibilities, which the computer program would avoid to maintain the most stable structure it could throughout the process. Human gamers are also known to have pattern-recognition skills and puzzle-solving intuition. Couple these traits with some creativity and natural human curiosity, and you get another step further towards understanding how proteins fold.
Structural biologists are driven by the idea that “structure equals function.” It means that knowing a protein’s structure allows us to understand how it functions. If proteins play a role in every single living organism, knowing how each one works is powerful knowledge. While this set of player-designed structures had no biological purpose, new designs could. “Since proteins are part of so many diseases, they can also be part of the cure. Players can design brand new proteins that could help prevent or treat important diseases,” say FoldIt's creators. Isn’t this one step closer?



Nice article Luyi! As a science communicator, I love to see how citizen science projects promote interaction between science and society. It is so cool to see how lay/non-scientist people can contribute to generate new science knowledge!