Rare spindle-shaped neurons from deep inside the brain recorded for the first time
Losing the mysterious cells may lead to Alzheimer's, schizophrenia, or other neurological disorders
About five years ago, researchers from the Allen Institute for Brain Science in Seattle received a special donation: a piece of a live, rare brain tissue. It came from a very deep part of the brain neuroscientists usually can't access. The donated tissue contained a rare and mysterious type of brain cells called von Economo neurons (VENs) that are thought to be linked to social intelligence and several neurological diseases.
The tissue was a byproduct of a surgery to remove a brain tumor from a patient in her 60s. The location of the tissue turned out to be in one of the deepest layers of the frontoinsular cortex, which is one of the few places where these rare neurons are found in the human brain. “This was one of the extremely rare chances that we received this tissue from a donor that had a tumor being removed from quite a deep [brain] structure,” said Rebecca Hodge, who is the co-first author of the study, published in Nature Communications on March 3rd. Hodge and her colleagues became the first scientists to record electrical spikes from these neurons. Further studies they did on these cells gave them clues about the VENs’ identity and function in the human brain.

Constantin von Economo
Wikimedia
VENs are large, spindle-shaped neurons. They were first identified by the Ukrainian scientist Vladimir Betz more than a century ago. They were later named after the anatomist Constantin von Economo, who described their shape and distribution through the human cortex. Only humans and especially social animals with large brains, such as great apes, whales, dolphins, and elephants have VENs. It is hypothesized that the cells evolved independently in these animals. Since common lab animals with smaller brains, like mice and rats, don't have VENs, it is difficult to study them in a lab environment.
Past studies linked VENs to social engagement and cognitive health. An analysis of "SuperAger" brains from older people who don’t suffer from the memory loss of "normal" aging showed a greater number of VENs compared to their cognitively average-for-their-age peers. Loss of VENs, on the other hand, has been observed in brains of patients suffering from a neurodegenerative disease called behavioral variant frontotemporal dementia, as well as from several other neurological disorders, including schizophrenia, autism, and possibly Alzheimer’s disease. None of these studies, however, offered clues about VENs’ exact function or unique properties. Unraveling the mystery of these neurons can help find therapies for these disorders.
The team at the Allen Institute tackled this mystery with two parallel studies: The first aimed at understanding VENs’ electrical properties, while the second one focused on their genetic identity. They didn't go exactly as planned.
For the first study, neuroscientists Brian Kalmbach and Jonathan Ting, from the Allen Institute decided to capture VENs’ electrical activity using method called patch clamp. Patch clamp is a very delicate technique where a scientist carefully punctures a cell with a very thin piece of glass to record its electrical activity.
The scientists were experienced with this technique, but the VENs they received were much more fragile than what they were used to. Even a gentle touch would be enough to make them explode. In the end, the scientists were able to record from only three neurons. The neurons showed unique electrical properties compared to other neuron types. Though the sample size was small, it was still the first ever recording of VENs, and the data was promising.
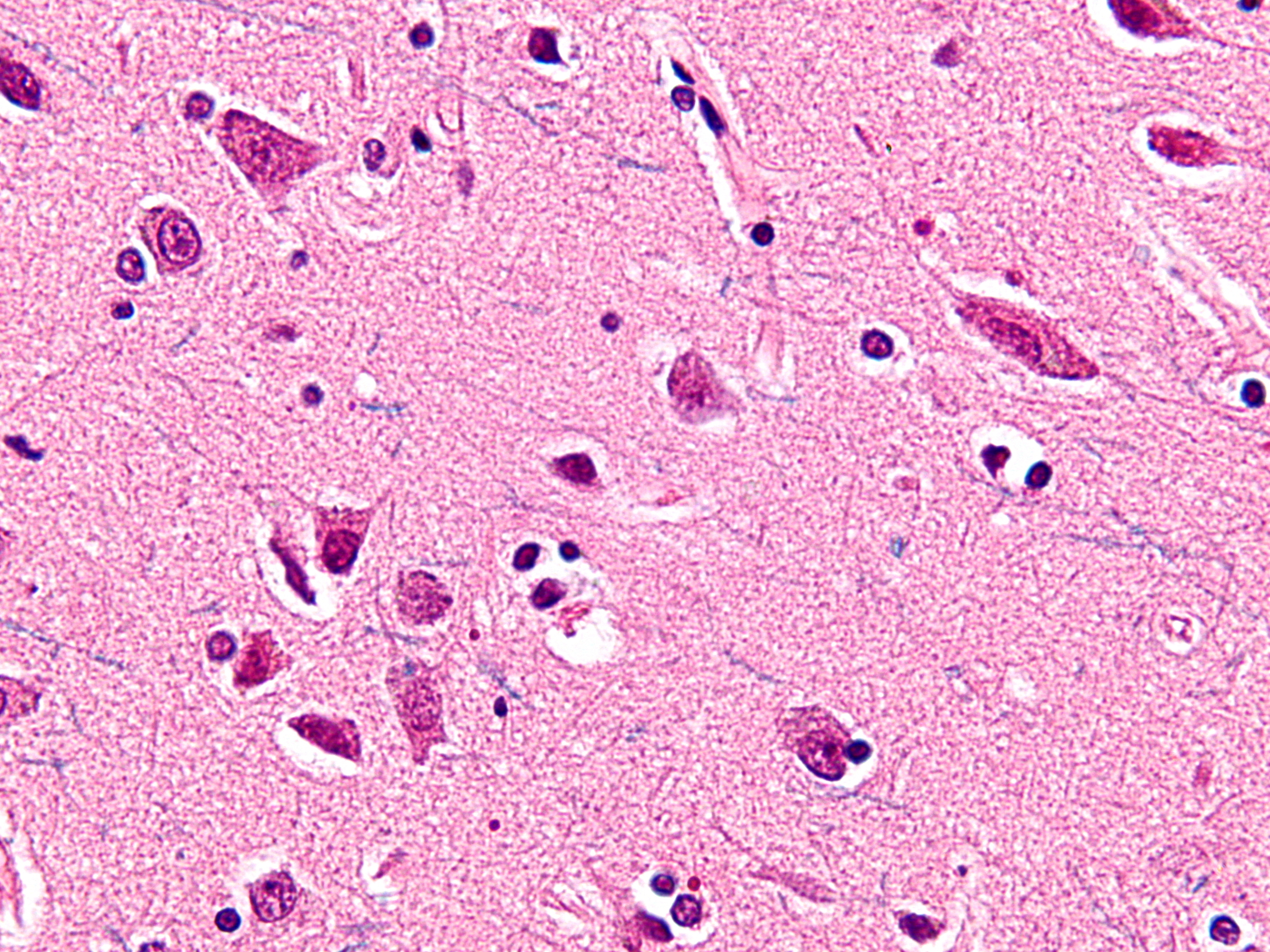
Spindle neurons
Wikimedia
The second study provided more answers. Hodge and colleagues wanted to genetically identify VENs using a new genome sequencing technique they were trying to develop at the time called single nucleus RNA-sequencing. “We had the goal of turning that technique into kind of a big data pipeline,” Hodge said. "but we didn't have any methods for doing that in humans yet."
The question was: how do VENs genetically differ from the other neurons in the same region?
One of the challenges with this study was how sparse VENs are in the human brain. They only account for a very small fraction, about 1.25 percent, of all neurons in the frontoinsular cortex, and about 500,000 neurons brain-wide. The group was only able to capture data from a handful of them. “So that's sort of a big challenge of working in human brain where you have you don't have nice transgenic tools like you have in mouse,” Hodge said. "You just have to see what you can get using the tools that you have available."
From the gene sequencing analysis the group was able to identify new marker genes for VENs that could be used to differentiate them from other neurons besides looking at their unique shape (which isn't always easy due to their rarity). But, the data set they got from the study was also too small to interpret the results accurately.
To tackle this problem, they compared these human cells to cell types that were already defined in mouse tissue to see if any of them matched using a computational mapping technique. Jeremy Miller, the other co-first author of the paper and the bioinformatician that performed the analysis, called the technique “a computational advance” that allowed them to predict what kind of class of cells VENs belonged to. The finding was surprising. Given the unique shape of VENs, the scientists expected them not to match with any other cell types, but they did. VENs’ genetic signatures looked very similar to those of neurons that send their axons from the cortex to deeper regions of the brain called extratelencephalic-projecting (ET) excitatory neurons.
Miller thought this finding could have interesting implications for neurodegenerative diseases, such as the behavioral variant frontotemporal dementia, where VENs are thought to be selectively vulnerable. Future studies might look to see whether it is all of the ET-type neurons that are lost in disease or whether it's only VENs. This information would help with deciding which cell types to target for therapies. The group is currently repeating the study using new sequencing methods that allow them to get tens of thousands to millions of cells in order to better understand the genetic properties of these neurons.






This is the first I’m hearing of these neurons and your article has made me want to find out more about them. Given how difficult and rare it is for scientists to get a sample of VENs, did they try to culture them to get a sustainable source with higher numbers? I’m also curious about the similarity in the transcriptome of human VENs and mice ETs. Have there been studies to see if mice ETs have a subset which act as their VENs? Did the authors also compare their electrophysiology to see if they behaved similarly? That would open up a ton of possibilities.