.jpg?auto=compress%2Cformat&crop=faces&fit=crop&fm=jpg&h=360&q=70&w=540
)
HIV uses camouflage to hide from cell defenses
Viruses are fighting an evolutionary arms race in our DNA
In 2000, Bill Clinton announced the creation of, “the most wondrous map ever produced by human kind,” a complete sequence of the human genome. During the two decades since, biologists have merrily sequenced the DNA of hundreds of animals, delighting in the plethora of data. The discoveries have illuminated many biological mysteries, including which gene sequences play roles in diseases, and helped sketch out evolutionary family trees. But so much data has also generated entirely new biological mysteries – weird patterns and sequence oddities that still puzzle scientists.
DNA is a long string of four unique molecules, called nucleotides. Each nucleotide molecule goes by a shorthand, one-letter abbreviation of its chemical name: A, G, C, or T. The combinations of these nucleotides form an inheritable biological code. When biologists first began to explore the sequences of nucleotides, they noticed something strange: vertebrate genomes have very few of a particular combination of two DNA molecules, C and G nucleotides, in that order. Why the missing CG pairs? It’s thought that the dearth of CGs is due to unique chemical properties of the combination, which predispose mutations that swap the C nucleotide for a T. Over millions of years, these accumulated mutations have dramatically reduced the CG content in our DNA.
Vertebrates aren't the only lifeforms to lack CG pairs. Although HIV, and viruses like it, have genomes made of RNA, a close chemical cousin to DNA, they also have a scarcity of CG pairs. But the chemical properties that induce mutations in CG pairs in DNA don’t exist in RNA, meaning the viruses must lack CG pairs for another reason. Until recently, the lack of CGs in HIV has been a thorny puzzle for virologists.
In October, biologists at Rockefeller University shed light on the mystery: they discovered that viruses have taken advantage of this unique vertebrate DNA fingerprint in order to camouflage themselves in their hosts. When scientists destroyed the camouflage, by adding back CG nucleotide pairs into the genomes of HIV viruses, host cells were able to mount effective defenses, and dramatically reduce the number of viruses.
In their research, the scientists went on to describe a particular protein within host cells that scan for CG pairs, and destroy the DNA or RNA when they find it. HIV that contain very few CG pairs survive the surveillance, but HIV with many CG pairs are eradicated. The authors proposed that HIV has evolved to evade this type of host detection by reducing the number of CG pairs in its genome.
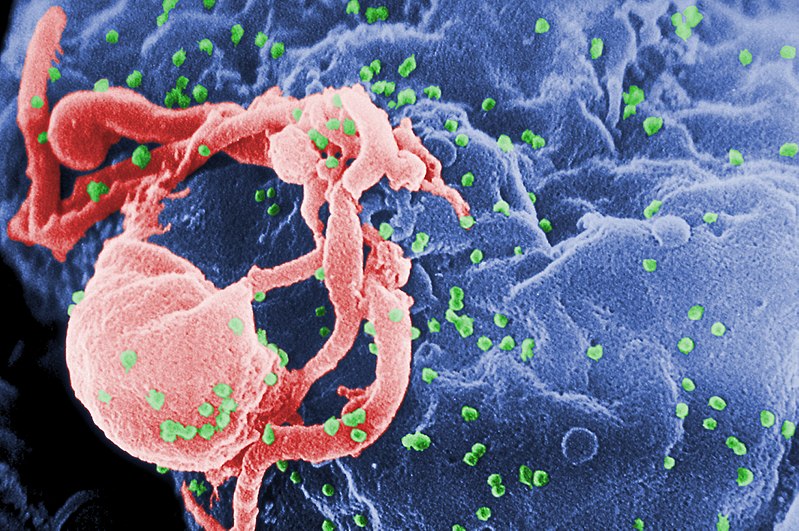
HIV, highlighted green
When a virus infects a human cell, it hijacks cellular machinery to churn out more virus components, effectively building a self-replicating factory. To do this, the virus must subvert a lot of security systems within the host cell, which have been optimized over the course of evolutionary history to detect invading viruses and destroy them before too much damage is done.
Subcellular scavenger hunts
The biologists at Rockefeller discovered an entirely new security system in our cells. Swimming in the cytoplasm of vertebrate cells are proteins that, on finding molecules with CG nucleotide pairs, make chemical bonds specifically with those CG pairs – and then destroy the molecules. Since human cells don’t have many CGs in their DNA, any pairs detected in the cell are probably from a virus or bacteria – an interloper swiftly degraded by cell defenses. This seems like a great survival strategy, but unfortunately, we’ve been outsmarted by the viruses, which have co-opted our unique DNA fingerprint to camouflage themselves.
It’s an evolutionary arms race. Vertebrate cells, through an accident of chemistry, happen to have few CG nucleotide pairs. Because of this, any CG nucleotides floating around in a cell are probably from an outside intruder. Over millions of years, cells have evolved protein machinery that patrol for those CG pairs. In reaction, viruses like HIV evolved to have fewer and fewer CG pairs, with natural selection sculpting their genomes to match those of their hosts.
Serendipity played no small part in the discovery. Matthew Takata, the graduate student who authored the study, began his scientific career as a landlocked marine biologist, studying the cell biology of sea urchins while in Minnesota, thousands of miles from the sea. More than five years later later, he’s now wrapping up his PhD at Rockefeller University. “Before coming to Rockefeller, I was never interested in viruses,” he chuckled. But as a new student in New York City, Takata became enthralled by the work of Paul Bieniasz, a virologist whose lab he eventually joined.
Takata set off in search of sequences in the HIV genome that make it easier for human cells to mitigate infection. Human cells became better HIV-fighters, he noticed, when mutations in the virus increased CG pairs. Armed with the surprising observation, he started to look for human proteins that might be able to detect the viral CGs. Biologists often take on these subcellular scavenger hunts by tinkering with genes of interest, and observing what ensues in cells.
On the hunt, Takata altered many, many human genes, most of which didn’t have any effect on cells' ability to detect the viral CGs. At the very end of his search, just when he was considering switching experimental directions, he stumbled across a few papers that described a human gene called ZAP. ZAP looked like it might fit the bill for the CG sensor protein – it was just about the right size and shape, and there was some evidence that the protein could bind to RNA. Incredibly, when Takata destroyed ZAP, and infected human cells with the CG-enriched viruses, the cells couldn’t mount an effective defense.
Takata went on to show that ZAP was, in fact, binding to the viral CG pairs, and he now hopes that the protein could inspire new antiviral drugs. Additionally, the discovery of CG camouflage could lead to new vaccine designs; if we were able to make vaccines with a CG-enriched virus, we could kick-start the host's immune system without exposing patients to more dangerous forms of the virus.
The research is a case study of how one puzzling observation – in this case about gene sequences in vertebrates – uncovered an evolutionary arms race, and a key cell defender that our bodies use to sniff out and destroy viruses. Armed with the discovery, we can use it to our advantage, designing drugs that bolster the native anti-viral defense systems.



This is a cool story of self/non-self discrimination based on stable GC vs AT content in the human genome. So there are really two selection processes for invading viruses to contend with: positive selection for the viral genes to be easily transcribed and translated (codon-usage that matches humans', for example), or negative selection against alien-looking GC content.
(Semi-unrelatedly,) in some bacteria there is the opposite phenomenon: foreign DNA that has too many AT pairs is toxic, and there are mechanisms to recognize and "silence" (prevent expression of genes in) the AT-rich invading DNA.