
Photo by Alexander Popov on Unsplash
DNA-RNA hybrid molecules keep the very centers of our cells together
Complex arrangements of these molecules keep the molecular machinery in our cells working properly
Our DNA molecules look like zippers: two strands of DNA hooked together. The information in the DNA is read and transcribed to make DNA’s alter ego – RNA. The information in RNA is then read by protein fabricators, called ribosomes, to make proteins.
DNA and RNA are like ropes that can be unzipped, stretched, looped, rolled, wound, knotted, and zipped back together. At times when DNA is unzipped, like when RNA synthesizing enzymes are reading the information in it, RNA can stick to one of the DNA strands like hook and loop tapes.
This combination of RNA stuck with DNA makes a bizarre structure called a DNA-RNA hybrid. These structures are stronger than DNA itself and are like knots in a usually long rope of DNA.
In the past two decades, many labs have reported that these hybrids are bad for DNA because cells spend more energy and resources to keep them in check. When cells accumulate hybrids, these hybrids can eventually challenge DNA's stability and even break the genome apart (a case often observed in cancer cells).
But a group of scientists based at the University of Toronto recently reported that DNA-RNA hybrids can be good when they form in a specific region of the DNA that does not contain information to make proteins (it is estimated that only about 1-2% of the information in our DNA codes for proteins).
This specific region is located between ribosomal genes (rDNA) that code for the ribosomal RNA molecules that make proteins. Unlike other genes, which are, for the most part, randomly scattered across the genome, rDNA genes are clustered in the same general location. There are large gaps between them that are filled with buffering DNA sequences (written as "gap DNA" for convenience). You can think of it like a freight train with covered cars carrying goods –the rDNA – separated by large flat, empty cars (the gap DNA).
In humans, one "rDNA unit" is made up of rDNA gene cluster and a large gap DNA. About seventy such units are present in each of five of the human chromosomes. They jumble together, looped and jam-packed, so much so that when we look at a human cell using a table-top microscope, we can see them as a dense, bustling spot inside the nucleus called the nucleolus.
This new study was born from an observation that Karan Abraham, one of the researchers in this study, made about six years ago. Using a super-resolution microscope, he observed that two different RNA synthesizing enzymes – RNAPI and RNAPII – cohabitate in the nucleolus.
These enzymes latch onto DNA and move across it, scanning information and printing RNA molecules along the way. While it is long known that RNAPI runs through the fields of rDNA genes making new rRNA molecules; the team found RNAPII latching exclusively at the gap DNA and working from the opposite direction toward RNAPI. Using the freight train analogy, it is as if two people are running towards each other on the train, with one (RNAPI) jumping across the covered cars and the other (RNAPII) on the flat open cars.
Now when they treated cells with specific chemicals to block RNAPII, they found that the rRNA production by RNAPI was also impeded, suggesting that RNAPII, though operating only the gap DNA, was somehow helping RNAPI do its job.
Interestingly, when they blocked RNAPII, RNAPI went wild and hopped into the gap DNA: this stressed out the entire cell. The nucleolus, unlike most cell organelles, is not enclosed by a membrane but coalesces around rDNA clusters in the genome, similar to how vinegar separates in oil. The researchers saw that when RNAPI jumped into the gap DNA, the nucleolus sensed this as a form of cellular stress and started to fall apart.
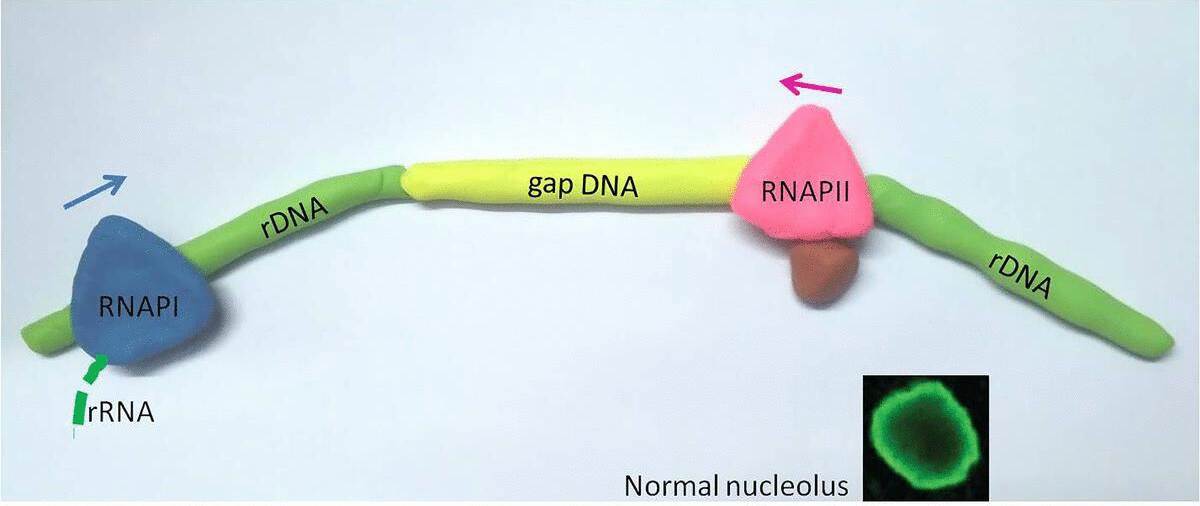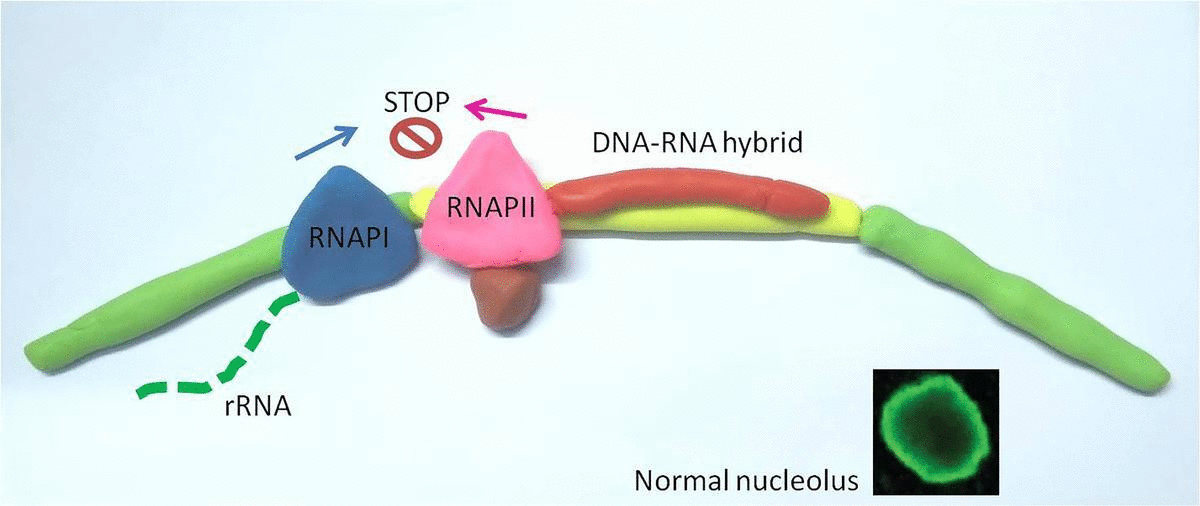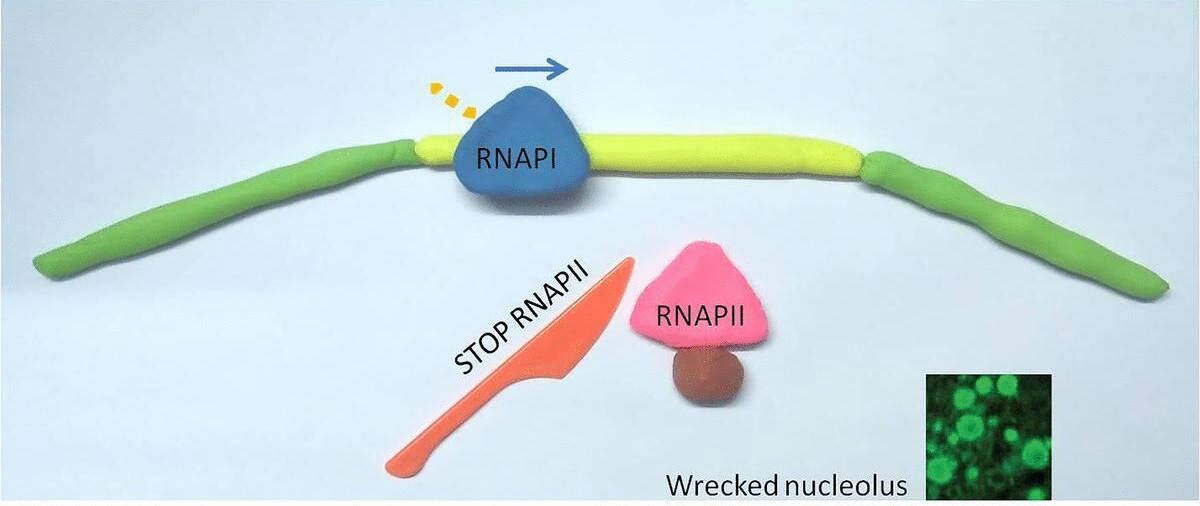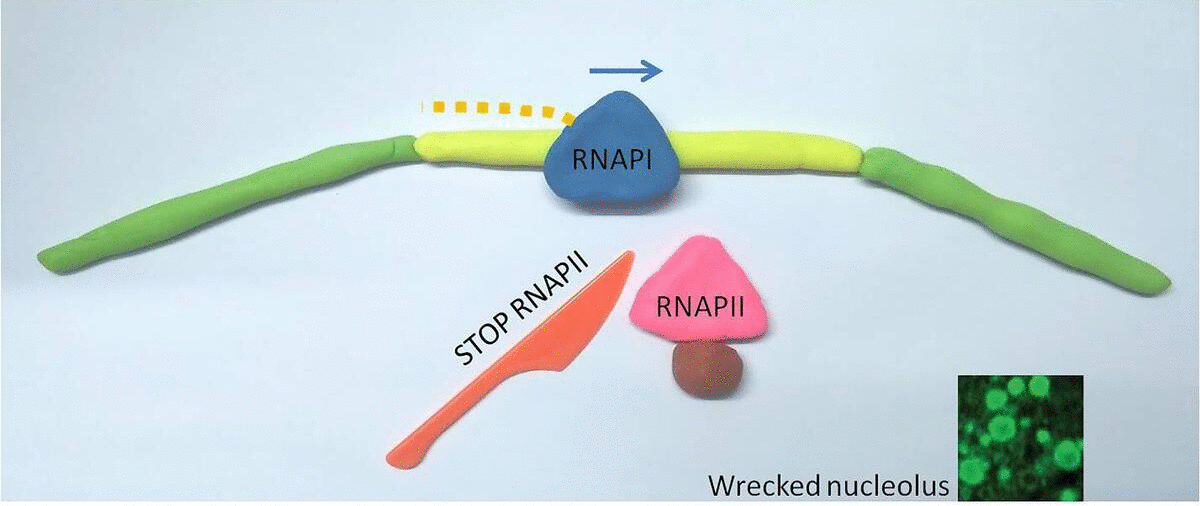
In a healthy cell, RNA Polymerase II (RNAPII) occupies the gaps between ribosomal genes, where RNA Polymerase I (RNAPI) resides. When RNAPII is blocked (represented by a red knife), RNAPI overruns it's normal genetic territory, and the cell becomes unhealthy.
Sree Rama Chaitanya
Based on this, the team hypothesized that RNAPII could be preventing the RNAPI from jumping into the gap DNA. This is where the DNA-RNA hybrids come into action – they found that RNAPII constructs a strong molecular "shield" with DNA-RNA hybrids at the gap DNA to stop RNAPI from jumping in.
rRNAs are the most abundant RNA molecules produced by a cell, accounting for 60-80 percent of RNA the cell makes. Having multiple copies of rDNA might not be sufficient to supply a cell's constant need for rRNAs. The rDNA must be kept together but not too close, and at a respectable distance to promote efficient production of rRNA at a regulated pace, for the same reason you wouldn't build multiple factories one on top of the other. It is likely that the complex arrangement of rDNA and gap DNA, combined with the intricate balance between biomolecules, keeps molecular machinery from piling up and interfering with each other.
These findings are important because such a balance often goes haywire in pathological conditions. For example, Ewing sarcoma cells lack a balanced level of DNA-RNA hybrids. Although far-fetched at this moment, future work could build on this to design new ways to tackle therapeutic strategies in Ewing sarcoma, or even other cancers.
